7XSX
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49) | | 分子名称: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | 著者 | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | 登録日 | 2022-05-15 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSZ
 
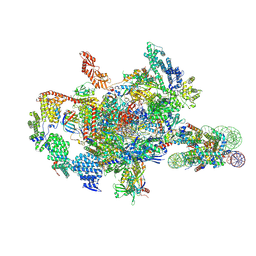 | | RNA polymerase II elongation complex transcribing a nucleosome (EC115) | | 分子名称: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | 著者 | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | 登録日 | 2022-05-15 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XT7
 
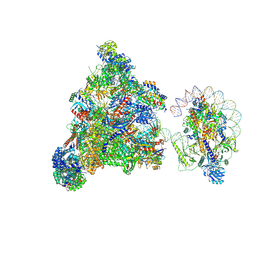 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49B) | | 分子名称: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | 著者 | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | 登録日 | 2022-05-16 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XTI
 
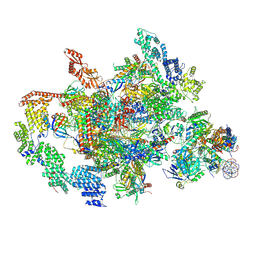 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58hex) | | 分子名称: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | 著者 | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | 登録日 | 2022-05-17 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
6M04
 
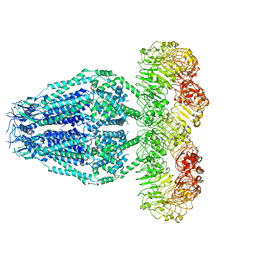 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | 分子名称: | Volume-regulated anion channel subunit LRRC8D | | 著者 | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | 登録日 | 2020-02-20 | | 公開日 | 2020-06-17 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
2PSM
 
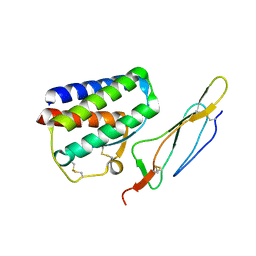 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | 分子名称: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | 著者 | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-05-07 | | 公開日 | 2007-11-06 | | 最終更新日 | 2021-08-18 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
6LM0
 
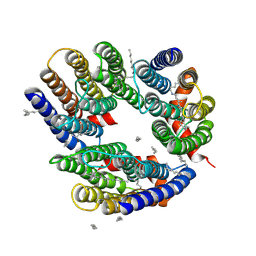 | | The crystal structure of cyanorhodopsin (CyR) N2098R from cyanobacteria Calothrix sp. NIES-2098 | | 分子名称: | DECANE, HEXANE, N-OCTANE, ... | | 著者 | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | 登録日 | 2019-12-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
6LM1
 
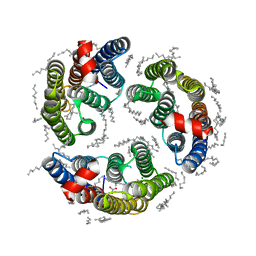 | | The crystal structure of cyanorhodopsin (CyR) N4075R from cyanobacteria Tolypothrix sp. NIES-4075 | | 分子名称: | DECANE, DODECANE, HEXADECANE, ... | | 著者 | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | 登録日 | 2019-12-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
7Y6L
 
 | |
7Y6N
 
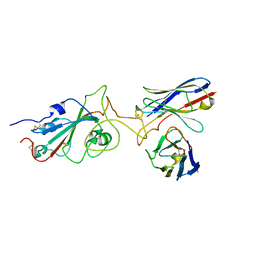 | |
1FJD
 
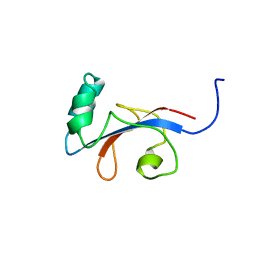 | | HUMAN PARVULIN-LIKE PEPTIDYL PROLYL CIS/TRANS ISOMERASE, HPAR14 | | 分子名称: | PEPTIDYL PROLYL CIS/TRANS ISOMERASE (PPIASE) | | 著者 | Terada, T, Shirouzu, M, Fukumori, Y, Fujimori, F, Ito, Y, Kigawa, T, Yokoyama, S, Uchida, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2000-08-08 | | 公開日 | 2001-08-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the human parvulin-like peptidyl prolyl cis/trans isomerase, hPar14.
J.Mol.Biol., 305, 2001
|
|
1GD8
 
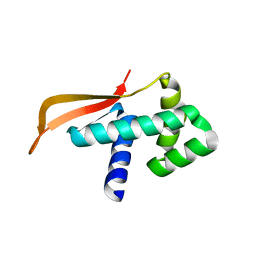 | |
1NZ8
 
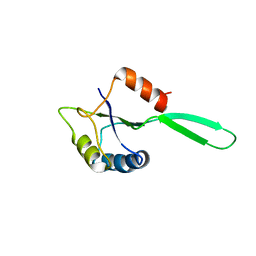 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | 分子名称: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | 著者 | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-02-17 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1PMS
 
 | | PLECKSTRIN HOMOLOGY DOMAIN OF SON OF SEVENLESS 1 (SOS1) WITH GLYCINE-SERINE ADDED TO THE N-TERMINUS, NMR, 20 STRUCTURES | | 分子名称: | SOS 1 | | 著者 | Koshiba, S, Kigawa, T, Kim, J, Shirouzu, M, Bowtell, D, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-02-18 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the pleckstrin homology domain of mouse Son-of-sevenless 1 (mSos1).
J.Mol.Biol., 269, 1997
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | 分子名称: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | 著者 | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-02-17 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
5GV0
 
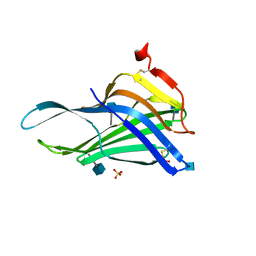 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | 著者 | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | 登録日 | 2016-09-01 | | 公開日 | 2016-10-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
5GV3
 
 | | Crystal structure of the membrane-distal domain of mouse lysosome-associated membrane protein 2 (LAMP-2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 2, ZINC ION | | 著者 | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | 登録日 | 2016-09-01 | | 公開日 | 2017-09-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.096 Å) | | 主引用文献 | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes.
Biochem. Biophys. Res. Commun., 479, 2016
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | 分子名称: | Splicing factor 3B subunit 4 | | 著者 | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2016-09-06 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
4LM5
 
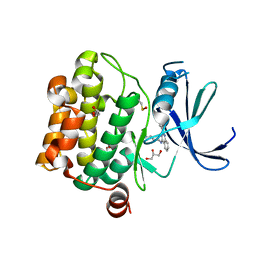 | | Crystal structure of Pim1 in complex with 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol (resulting from displacement of SKF86002) | | 分子名称: | 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol, GLYCEROL, Serine/threonine-protein kinase pim-1 | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-10 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUD
 
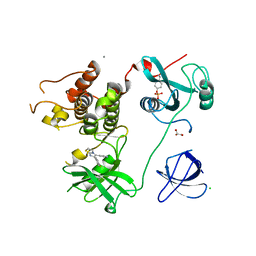 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | 分子名称: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-25 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUE
 
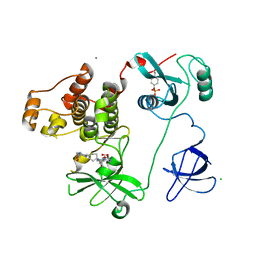 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | 分子名称: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-25 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LMU
 
 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | 分子名称: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-11 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | 分子名称: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-09 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5H09
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-ethyl2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoate | | 分子名称: | Tyrosine-protein kinase HCK, ethyl (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanoate | | 著者 | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | 登録日 | 2016-10-04 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.945 Å) | | 主引用文献 | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0B
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | 分子名称: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | 著者 | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | 登録日 | 2016-10-04 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
