5YZB
 
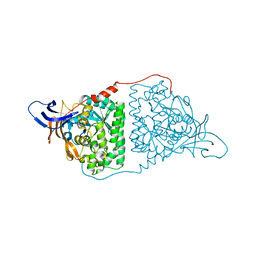 | | Crystal Structure of Human CRMP-2 with S522D-T509D-T514D-S518D mutations crystallized with GSK3b | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Imasaki, T, Sumi, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
5ZAB
 
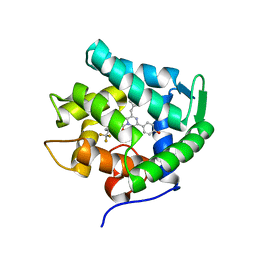 | | Crystal structure of cf3-aequorin | | Descriptor: | (2S)-8-benzyl-2-hydroperoxy-6-(4-hydroxyphenyl)-2-{[4-(trifluoromethyl)phenyl]methyl}imidazo[1,2-a]pyrazin-3(2H)-one, Aequorin-2 | | Authors: | Inouye, S, Tomabechi, Y, Sekine, S.I, Shirouzu, M, Hosoya, T. | | Deposit date: | 2018-02-07 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Slow luminescence kinetics of semi-synthetic aequorin: expression, purification and structure determination of cf3-aequorin.
J. Biochem., 164, 2018
|
|
5ZE9
 
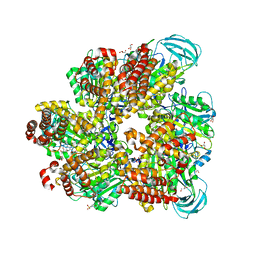 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5ZJ6
 
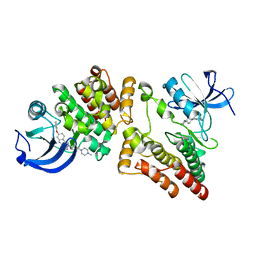 | | Crystal structure of HCK kinase complexed with a pyrrolo-pyrimidine inhibitor 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2018-03-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Phosphorylated and non-phosphorylated HCK kinase domains produced by cell-free protein expression.
Protein Expr. Purif., 150, 2018
|
|
5ZEA
 
 | | Crystal structure of the nucleotide-free mutant A3B3 | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.384 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
7C03
 
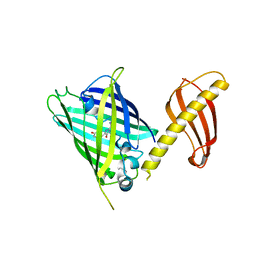 | | Crystal structure of POLArISact(T57S), genetically encoded probe for fluorescent polarization | | Descriptor: | POLArISact(T57S) | | Authors: | Tomabechi, Y, Sakai, N, Shirouzu, M. | | Deposit date: | 2020-04-30 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | POLArIS, a versatile probe for molecular orientation, revealed actin filaments associated with microtubule asters in early embryos.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8HU1
 
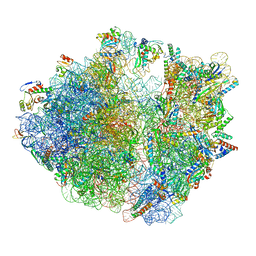 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and ct6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HTZ
 
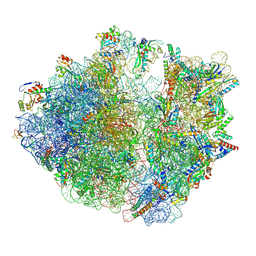 | | E. coli 70S ribosome complexed with H. marismortui tRNA_Ile2 bearing agm2C34 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7CLX
 
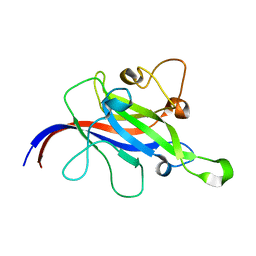 | | Crystal structure of the DOCK8 DHR-1 domain | | Descriptor: | Dedicator of cytokinesis protein 8 | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S, Fukui, Y, Uruno, T. | | Deposit date: | 2020-07-22 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A conserved PI(4,5)P2-binding domain is critical for immune regulatory function of DOCK8.
Life Sci Alliance, 4, 2021
|
|
7CLY
 
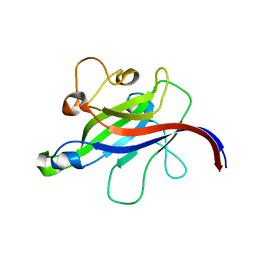 | | Structure of the DOCK8 DHR-1 domain crystallized with di-C8-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | Dedicator of cytokinesis protein 8 | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S, Fukui, Y, Uruno, T. | | Deposit date: | 2020-07-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | A conserved PI(4,5)P2-binding domain is critical for immune regulatory function of DOCK8.
Life Sci Alliance, 4, 2021
|
|
7C3G
 
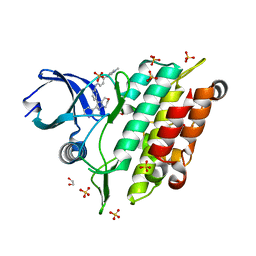 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
7CD1
 
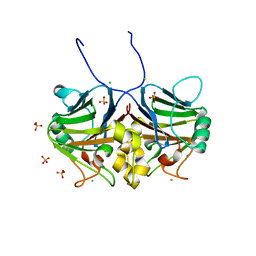 | | Crystal structure of inhibitory Smad, Smad7 | | Descriptor: | CHLORIDE ION, Mothers against decapentaplegic homolog 7, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for inhibitory effects of Smad7 on TGF-beta family signaling.
J.Struct.Biol., 212, 2020
|
|
7DDQ
 
 | | Structure of RC-LH1-PufX from Rhodobacter veldkampii | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2020-10-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-angstrom resolution.
Sci Adv, 7, 2021
|
|
7DPA
 
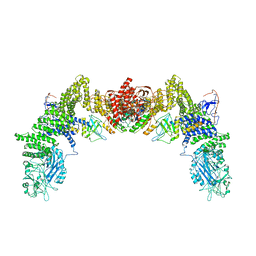 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
8HSP
 
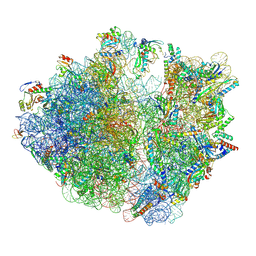 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and t6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-20 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7DEV
 
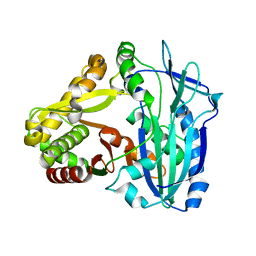 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora | | Descriptor: | Anthocyanin 5-aromatic acyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
7DEX
 
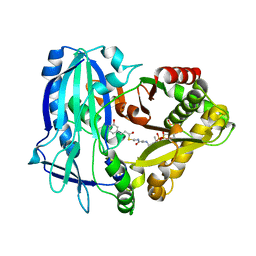 | | Crystal Structures of Anthocyanin 5,3'-aromatic acyltransferase H174A mutant with caffeoyl-CoA | | Descriptor: | Anthocyanin 5-aromatic acyltransferase, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (E)-3-[3,4-bis(oxidanyl)phenyl]prop-2-enethioate | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-11-05 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthocyanin 5,3'-aromatic acyltransferase from Gentiana triflora, a structural insight into biosynthesis of a blue anthocyanin.
Phytochemistry, 186, 2021
|
|
8KCQ
 
 | | Solution structures of the N-terminal divergent caplonin homology (NN-CH) domains of human intraflagellar transport protein 54 | | Descriptor: | TRAF3-interacting protein 1 | | Authors: | Dang, W, Kuwasako, K, He, F, Takahashi, M, Tsuda, K, Nagata, T, Tanaka, A, Kobayashi, N, Kigawa, T, Guentert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2023-08-08 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structure of the N-terminal divergent calponin homology (NN-CH) domain of human intraflagellar transport protein 54.
Biomol.Nmr Assign., 18, 2024
|
|
7DQE
 
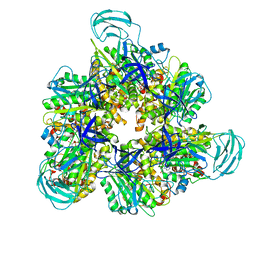 | | Crystal structure of the ADP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Maruyama, S, Nakamoto, K, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | The Combination of High-Speed AFM and X-ray Crystallography Reveals Rotary Catalytic Mechanism of Shaftless V1-ATPase
To Be Published
|
|
7DQD
 
 | | Crystal structure of the AMP-PNP-bound mutant A(S23C)3B(N64C)3 complex from enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, T. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.383 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
To Be Published
|
|
7DQC
 
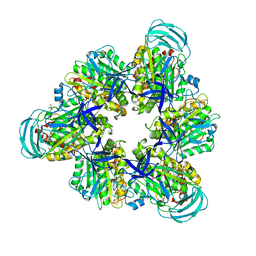 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
7EG2
 
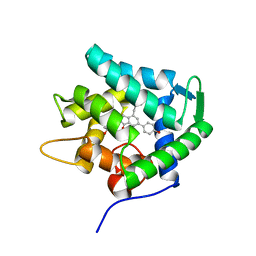 | | Crystal structure of the apoAequorin complex with (S)-daCTZ | | Descriptor: | (2~{S})-2-(hydroxymethyl)-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-3~{H}-inden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EG3
 
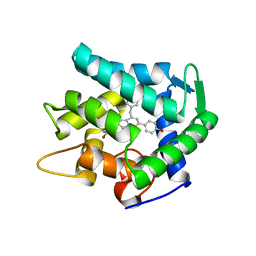 | | Crystal structure of the apoAequorin complex with (S)-HM-daCTZ | | Descriptor: | (2~{S})-6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-4-(phenylmethyl)-2,3-dihydroinden-1-one, Aequorin-2 | | Authors: | Tomabechi, Y, Shirouzu, M. | | Deposit date: | 2021-03-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Chiral deaza-coelenterazine analogs for probing a substrate-binding site in the Ca2+-binding photoprotein aequorin.
Plos One, 16, 2021
|
|
7EJL
 
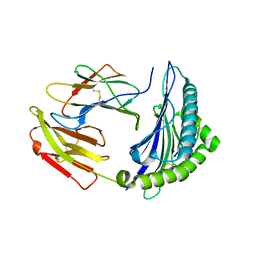 | |
7EJN
 
 | |
