5GKF
 
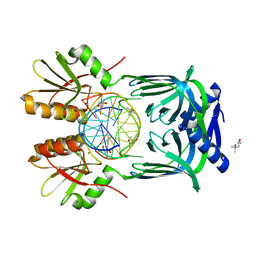 | | Structure of EndoMS-dsDNA1' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*TP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKE
 
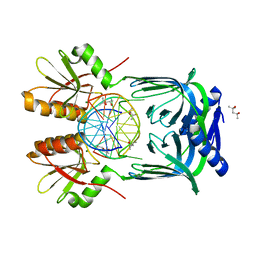 | | Structure of EndoMS-dsDNA1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*TP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
5GKH
 
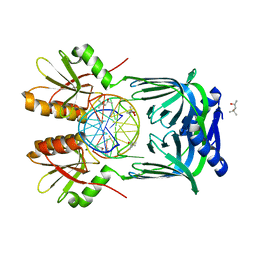 | | Structure of EndoMS-dsDNA2 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*CP*GP*GP*CP*AP*CP*TP*TP*GP*GP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*CP*CP*AP*GP*GP*TP*GP*CP*CP*GP*T)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
6IES
 
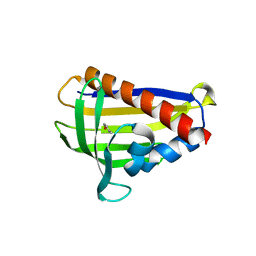 | | Onion lachrymatory factor synthase (LFS) containing (E)-2-propen 1-ol (crotyl alcohol) | | Descriptor: | (2E)-but-2-en-1-ol, Lachrymatory-factor synthase | | Authors: | Sato, Y, Arakawa, T, Takabe, J, Masamura, N, Tsuge, N, Imai, S, Fushinobu, S. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the Stereocontrolled Conversion of Short-Lived Sulfenic Acid by Lachrymatory Factor Synthase.
Acs Catalysis, 10, 2020
|
|
5GKG
 
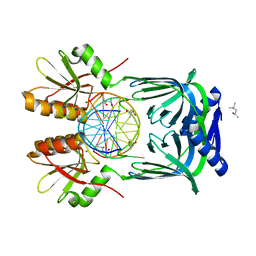 | | Structure of EndoMS-dsDNA1'' complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*CP*GP*CP*TP*AP*CP*AP*GP*GP*TP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*AP*CP*GP*TP*GP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Nakae, S, Hijikata, A, Tsuji, T, Yonezawa, K, Kouyama, K, Mayanagi, K, Ishino, S, Ishino, Y, Shirai, T. | | Deposit date: | 2016-07-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the EndoMS-DNA Complex as Mismatch Restriction Endonuclease
Structure, 24, 2016
|
|
7EXW
 
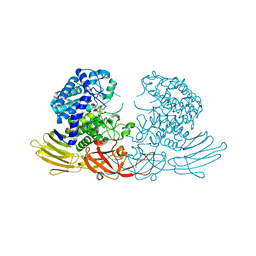 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | Descriptor: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXU
 
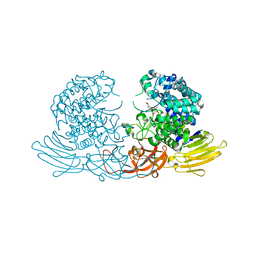 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | Descriptor: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | Descriptor: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
8CZ8
 
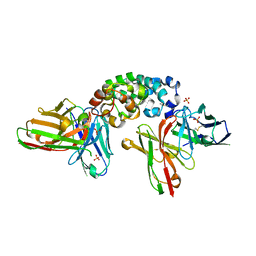 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
4UBS
 
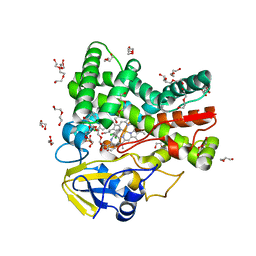 | | The crystal structure of cytochrome P450 105D7 from Streptomyces avermitilis in complex with Diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Xu, L.H, Ikeda, H, Arakawa, T, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
7YQS
 
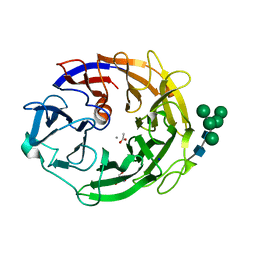 | | Neutron structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (1.25 Å), X-RAY DIFFRACTION | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
1IZ4
 
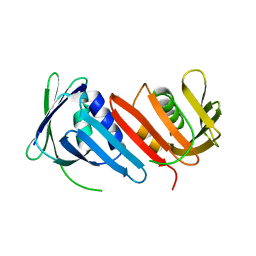 | |
1IZ5
 
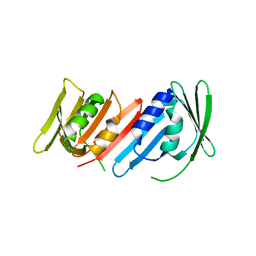 | | Pyrococcus furiosus PCNA mutant (Met73Leu, Asp143Ala, Asp147Ala): orthorhombic form | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-09-23 | | Release date: | 2003-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA
PROTEIN SCI., 12, 2003
|
|
8CXC
 
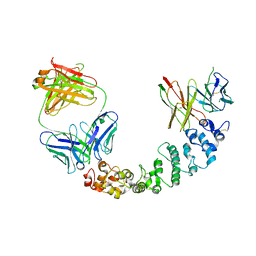 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
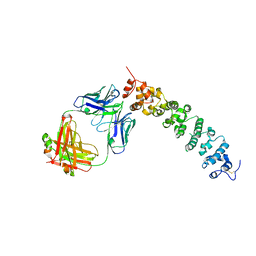 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
1ISQ
 
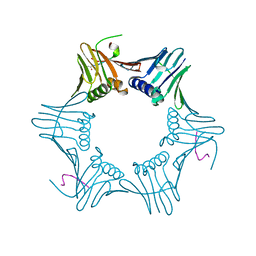 | | Pyrococcus furiosus PCNA complexed with RFCL PIP-box peptide | | Descriptor: | Proliferating Cell Nuclear Antigen, replication factor C large subunit | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2001-12-19 | | Release date: | 2002-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Physical interaction between proliferating cell nuclear antigen and
replication factor C from Pyrococcus furiosus
Genes Cells, 7, 2002
|
|
1IQP
 
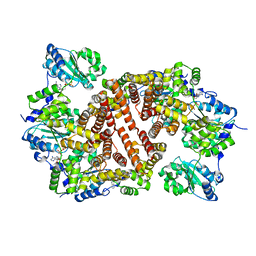 | | Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RFCS | | Authors: | Oyama, T, Ishino, Y, Cann, I.K.O, Ishino, S, Morikawa, K. | | Deposit date: | 2001-07-24 | | Release date: | 2001-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus
Mol.Cell, 8, 2001
|
|
7V1W
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-arabinofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase (alphaFFase1), beta-D-arabinofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1X
 
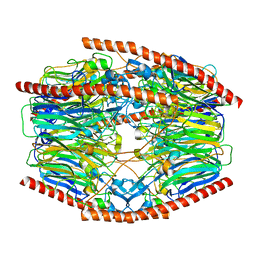 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium in complex with beta-D-fructofuranose | | Descriptor: | CALCIUM ION, Difructose dianhydride I synthase/hydrolase, beta-D-fructofuranose | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
7V1V
 
 | | Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, D(-)-TARTARIC ACID, ... | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | Deposit date: | 2021-08-06 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family.
J.Biol.Chem., 297, 2021
|
|
8WOP
 
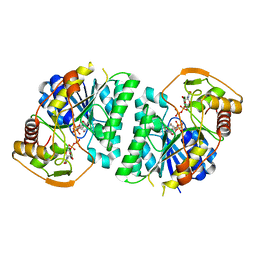 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, wild-type | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8WOW
 
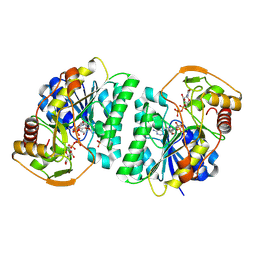 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, I160L/G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, UDP-glucose 4-epimerase 2, ... | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8WUC
 
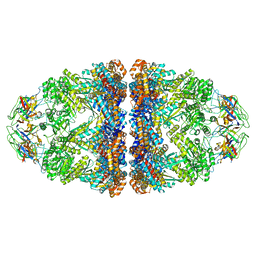 | | Cryo-EM structure of H. thermoluteolus GroEL-GroES2 football complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-20 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8WUW
 
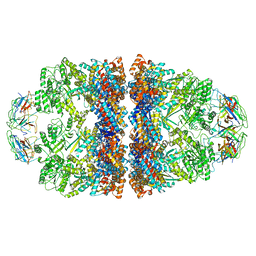 | | Cryo-EM structure of H. thermophilus GroEL-GroES2 asymmetric football complex | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, MAGNESIUM ION, ... | | Authors: | Liao, Z, Gopalasingam, C.C, Kameya, M, Gerle, C, Shigematsu, H, Ishii, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2023-10-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into thermophilic chaperonin complexes.
Structure, 32, 2024
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
