2RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
3VOV
 
 | | Crystal Structure of ROK Hexokinase from Thermus thermophilus | | Descriptor: | GLYCEROL, Glucokinase, ZINC ION | | Authors: | Nakamura, T, Kashima, Y, Mine, S, Oku, T, Uegaki, K. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and crystal structure of the thermophilic ROK hexokinase from Thermus thermophilus
J.Biosci.Bioeng., 2012
|
|
7Q5X
 
 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH 2-OXOGLUTARATE (2OG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
7W9Q
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-14 | | Descriptor: | (2~{R})-2-(3-chloranyl-4-oxidanyl-phenyl)-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, CALCIUM ION, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
7W9R
 
 | | Crystal structure of V30M-TTR in complex with naringenin derivative-18 | | Descriptor: | (2~{R})-2-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Transthyretin | | Authors: | Katayama, W, Shimane, A, Nabeshima, Y, Yokoyama, T, Mizuguchi, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Chlorinated Naringenin Analogues as Potential Inhibitors of Transthyretin Amyloidogenesis.
J.Med.Chem., 65, 2022
|
|
7Q5V
 
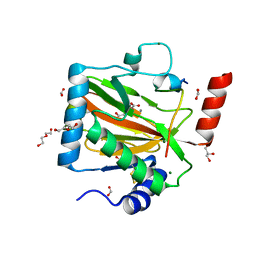 | | HIF PROLYL HYDROXYLASE 2 (PHD2/EGLN1) IN COMPLEX WITH N-OXALYLGLYCINE (NOG) AND HIF-2 ALPHA CODD (523-542) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Chowdhury, R, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural basis for binding of the renal carcinoma target hypoxia-inducible factor 2 alpha to prolyl hydroxylase domain 2.
Proteins, 91, 2023
|
|
2MLQ
 
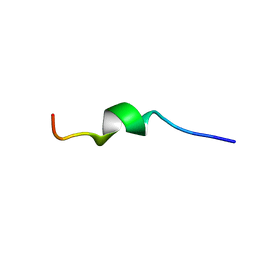 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
1A0A
 
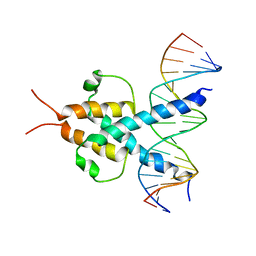 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
1EWV
 
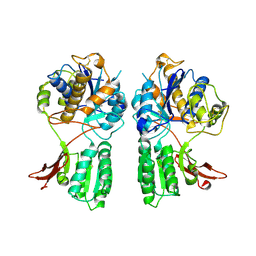 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
1EWK
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
7FFC
 
 | | Diarylpentanoid-producing polyketide synthase (A210E mutant) | | Descriptor: | GLYCEROL, Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
5X9S
 
 | | Crystal structure of fully modified H-Ras-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Ke, H, Murashima, Y, Taniguchi-Tamura, H, Miyamoto, R, Yoshikawa, Y, Kumasaka, T, Mizohata, E, Edamatsu, H, Kataoka, T. | | Deposit date: | 2017-03-09 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for intramolecular interaction of post-translationally modified H-RasGTP prepared by protein ligation
FEBS Lett., 591, 2017
|
|
5H0Q
 
 | | Crystal structure of lipid binding protein Nakanori at 1.5A | | Descriptor: | Lipid binding protein | | Authors: | Makino, A, Abe, M, Ishitsuka, R, Murate, M, Kishimoto, T, Sakai, S, Hullin-Matsuda, F, Shimada, Y, Inaba, T, Miyatake, H, Tanaka, H, Kurahashi, A, Pack, C.G, Kasai, R.S, Kubo, S, Schieber, N.L, Dohmae, N, Tochio, N, Hagiwara, K, Sasaki, Y, Aida, Y, Fujimori, F, Kigawa, T, Nishikori, K, Parton, R.G, Kusumi, A, Sako, Y, Anderluh, G, Yamashita, M, Kobayashi, T, Greimel, P, Kobayashi, T. | | Deposit date: | 2016-10-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C
FASEB J., 31, 2017
|
|
6ZBO
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in Complex with 1-(6-morpholinopyrimidin-4-yl)-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-5-ol (Molidustat) | | Descriptor: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)pyrazol-3-ol, CHLORIDE ION, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
6ZBN
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in complex with tert-butyl 6-(5-hydroxy-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-1-yl)nicotinate (IOX4) | | Descriptor: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
8H8Q
 
 | | Fab-amyloid beta fragment complex at neutral pH | | Descriptor: | CHLORIDE ION, Fab, GLN-LYS-CYS-VAL-PHE-PHE-ALA-GLU-ASP-VAL-GLY-SER-ASN-CYS-GLY, ... | | Authors: | Kita, A, Irie, K, Irie, Y, Matsushima, Y, Miki, K. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fab-amyloid beta fragment complex at neutral pH
To Be Published
|
|
4TWW
 
 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
7FFI
 
 | | Diarylpentanoid-producing polyketide synthase (F340W mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFG
 
 | | Diarylpentanoid-producing polyketide synthase (N199F mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFA
 
 | | Diarylpentanoid-producing polyketide synthase from Aquilaria sinensis | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFH
 
 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
6IN0
 
 | | Crystal structure of EphA3 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Ephrin type-A receptor 3 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IMZ
 
 | | Crystal structure of MTH1 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
