1J05
 
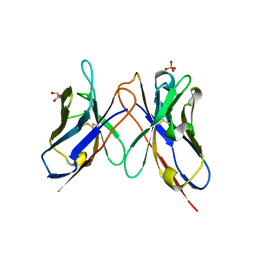 | | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment | | Descriptor: | GLYCEROL, PHOSPHATE ION, anti-CEA mAb T84.66, ... | | Authors: | Kondo, H, Nishimura, Y, Shiroishi, M, Asano, R, Noro, N, Tsumoto, K, Kumagai, I. | | Deposit date: | 2002-11-01 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment
To be Published
|
|
1UA6
 
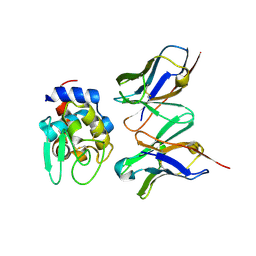 | | Crystal structure of HYHEL-10 FV MUTANT SFSF complexed with HEN EGG WHITE LYSOZYME complex | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-02-28 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.Biol.Chem., 278, 2003
|
|
7DTI
 
 | |
7DTH
 
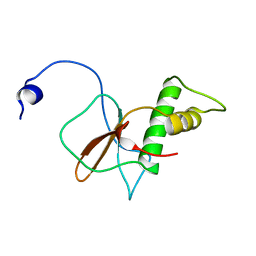 | | Solution structure of RPB6, common subunit of RNA polymerases I, II, and III | | Descriptor: | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | | Authors: | Okuda, M, Nishimura, Y. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three human RNA polymerases interact with TFIIH via a common RPB6 subunit.
Nucleic Acids Res., 50, 2022
|
|
1V29
 
 | | Crystal structure of Nitrile hydratase from a thermophile Bacillus smithii | | Descriptor: | COBALT (II) ION, nitrile hydratase a chain, nitrile hydratase b chain | | Authors: | Hourai, S, Miki, M, Takashima, Y, Mitsuda, S, Yanagi, K. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of nitrile hydratase from a thermophilic Bacillus smithii
Biochem.Biophys.Res.Commun., 312, 2003
|
|
1D8K
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1UAC
 
 | | Crystal Structure of HYHEL-10 FV MUTANT SFSF Complexed with TURKEY WHITE LYSOZYME | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Kumagai, I, Nishimiya, Y, Kondo, H, Tsumoto, K. | | Deposit date: | 2003-03-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10
J.BIOL.CHEM., 278, 2003
|
|
6JK5
 
 | | Ca2+-dependent type II antifreeze protein (Ca2+-free form) | | Descriptor: | SULFATE ION, Type II antifreeze protein | | Authors: | Arai, T, Tsuda, S, Kondo, H, Nishimiya, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Calcium-Binding Generates the Semi-Clathrate Waters on a Type II Antifreeze Protein to Adsorb onto an Ice Crystal Surface.
Biomolecules, 9, 2019
|
|
6JK4
 
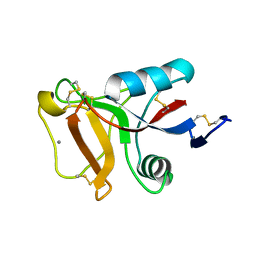 | | Ca2+-dependent type II antifreeze protein | | Descriptor: | CALCIUM ION, Type II antifreeze protein | | Authors: | Arai, T, Tsuda, S, Kondo, H, Nishimiya, Y. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Calcium-Binding Generates the Semi-Clathrate Waters on a Type II Antifreeze Protein to Adsorb onto an Ice Crystal Surface.
Biomolecules, 9, 2019
|
|
5B66
 
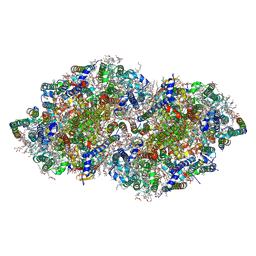 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
5B5E
 
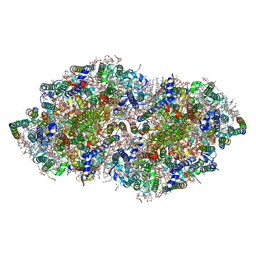 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
6IN2
 
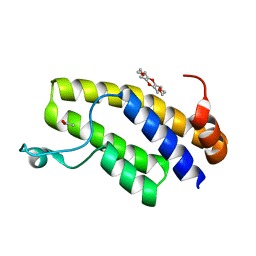 | | Crystal structure of BRD1 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ACETATE ION, Bromodomain-containing protein 1 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN1
 
 | | Crystal structure of the first bromodomain of BRD4 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN0
 
 | | Crystal structure of EphA3 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CHLORIDE ION, Ephrin type-A receptor 3 | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IMZ
 
 | | Crystal structure of MTH1 in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN3
 
 | | Crystal structure of DOT1L in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
6IN4
 
 | | Crystal structure of apo DAPK1 in the presence of 18-crown-6 | | Descriptor: | Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
1JL8
 
 | | Complex of alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with beta-cyclodextrin based on a co-crystallization with methyl beta-cyclodextrin | | Descriptor: | ALPHA-AMYLASE II, Cycloheptakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1WZK
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZM
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469K | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1JF6
 
 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
1JIB
 
 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
1D8J
 
 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1JF5
 
 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
