6WGE
 
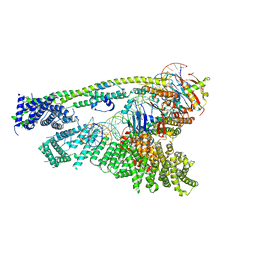 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | | 分子名称: | DNA (43-MER), Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | 著者 | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | 登録日 | 2020-04-05 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
7W1M
 
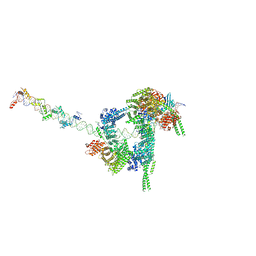 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | 著者 | Shi, Z.B, Bai, X.C, Yu, H. | | 登録日 | 2021-11-19 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
6WG3
 
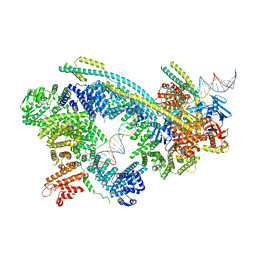 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | 分子名称: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | 著者 | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | 登録日 | 2020-04-04 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6WG6
 
 | |
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | 分子名称: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | 分子名称: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
6WG4
 
 | |
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | 分子名称: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4Z8M
 
 | | Crystal structure of the MAVS-TRAF6 complex | | 分子名称: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | 著者 | Shi, Z.B, Zhou, Z. | | 登録日 | 2015-04-09 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | 分子名称: | NB_1B11, Spike protein S1, ZINC ION | | 著者 | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | 登録日 | 2021-07-07 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
3CDJ
 
 | | Crystal structure of the E. coli KH/S1 domain truncated PNPase | | 分子名称: | Polynucleotide phosphorylase | | 著者 | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | 登録日 | 2008-02-27 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
3CDI
 
 | | Crystal structure of E. coli PNPase | | 分子名称: | Polynucleotide phosphorylase | | 著者 | Shi, Z, Yang, W.Z, Lin-Chao, S, Chak, K.F, Yuan, H.S. | | 登録日 | 2008-02-27 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation.
Rna, 14, 2008
|
|
5XY9
 
 | | Structure of the MST4 and 14-3-3 complex | | 分子名称: | 14-3-3 protein zeta/delta, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2017-07-06 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Structure of the MST4 and 14-3-3 complex
To Be Published
|
|
8Y3M
 
 | | Cryo-EM structure of DSR2-DSAD1 complex (cross-linked) | | 分子名称: | DSR anti-defence 1, SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3W
 
 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (same side) | | 分子名称: | DSR anti-defence 1, SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y13
 
 | | Cryo-EM structure of anti-phage defense associated DSR2 tetramer (H171A) | | 分子名称: | SIR2-like domain-containing protein | | 著者 | Li, F.X, Shi, Z.B, Wang, R.W, Xu, Q, Yang, R, Wu, Z.X. | | 登録日 | 2024-01-23 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8ZC9
 
 | | The Cryo-EM structure of DSR2-Tail tube-NAD+ complex | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein, tail tube protein | | 著者 | Wang, R, Xu, Q, Wu, Z, Li, J, Yang, R, Shi, Z, Li, F. | | 登録日 | 2024-04-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
2AOA
 
 | | Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain | | 分子名称: | 2-(4-((9S,10S,14S,Z)-18-(2-AMINO-2-OXOETHYL)-9-(CARBOXYMETHYL)-14-(NAPHTHALEN-1-YLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL)PHENYL)MALONIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Growth factor receptor-bound protein 2 | | 著者 | Phan, J, Shi, Z.D, Burke, T.R, Waugh, D.S. | | 登録日 | 2005-08-12 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal Structures of a High-affinity Macrocyclic Peptide Mimetic in Complex with the Grb2 SH2 Domain.
J.Mol.Biol., 353, 2005
|
|
2AXC
 
 | | Crystal structure of ColE7 translocation domain | | 分子名称: | Colicin E7, GLYCEROL, SULFATE ION | | 著者 | Cheng, Y.S, Shi, Z, Doudeva, L.G, Yang, W.Z, Chak, K.F, Yuan, H.S. | | 登録日 | 2005-09-04 | | 公開日 | 2006-03-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes
J.Mol.Biol., 356, 2006
|
|
8Y3Y
 
 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (opposite side) | | 分子名称: | DSR anti-defence 1, SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-29 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y34
 
 | | Cryo-EM structure of anti-phage defense associated DSR2 (H171A) (map2) | | 分子名称: | SIR2-like domain-containing protein | | 著者 | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | 登録日 | 2024-01-28 | | 公開日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
4KGB
 
 | |
4FI9
 
 | | Structure of human SUN-KASH complex | | 分子名称: | Nesprin-2, SUN domain-containing protein 2 | | 著者 | Wang, W.J, Shi, Z.B. | | 登録日 | 2012-06-08 | | 公開日 | 2012-07-18 | | 最終更新日 | 2013-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
4HK1
 
 | |
5GZB
 
 | | Crystal Structure of Transcription Factor TEAD4 in Complex with M-CAT DNA | | 分子名称: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*TP*TP*CP*CP*TP*CP*TP*C)-3'), GLYCEROL, ... | | 著者 | He, F, Shi, Z.B, Zhou, Z.C. | | 登録日 | 2016-09-28 | | 公開日 | 2017-04-19 | | 最終更新日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | DNA-binding mechanism of the Hippo pathway transcription factor TEAD4
Oncogene, 36, 2017
|
|
