5YK9
 
 | |
3AP3
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Protein-tyrosine sulfotransferase 2 | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
3AP1
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
2E5E
 
 | | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Matsumoto, S, Yoshida, T, Yasumatsu, I, Yamamoto, H, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts
to be published
|
|
3AP2
 
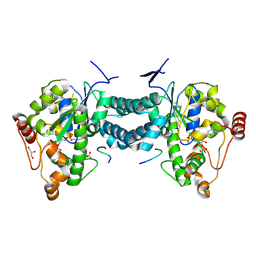 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP,C4 peptide, and phosphate ion | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2: Insights into substrate-binding and catalysis of post-translational protein tyrosine sulfation
To be Published
|
|
5Y4G
 
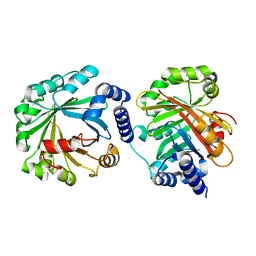 | | Apo Structure of AmbP3 | | Descriptor: | AmbP3 | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-03 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5X17
 
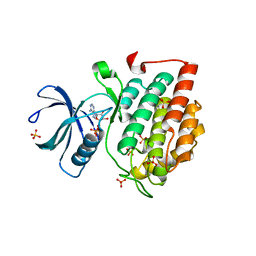 | | Crystal structure of murine CK1d in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, SULFATE ION | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
5Y72
 
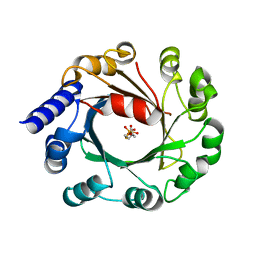 | | DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y84
 
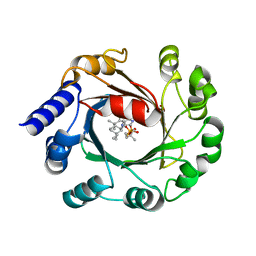 | | Hapalindole U and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole U | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y7C
 
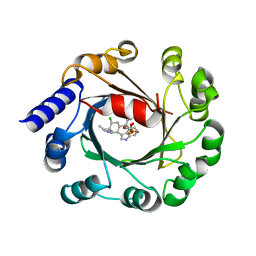 | | Hapalindole A and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole A | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5VOI
 
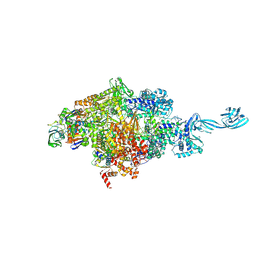 | | X-ray crystal structure of bacterial RNA polymerase and pyrG promoter complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murakami, K.S, Shin, Y, Turnbough Jr, C.L, Molodtsov, V. | | Deposit date: | 2017-05-02 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystal structure of a reiterative transcription complex reveals an atypical RNA extension pathway.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Z43
 
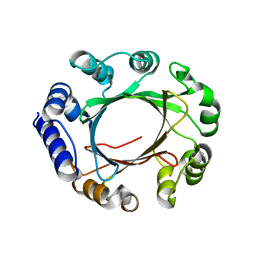 | | Crystal structure of prenyltransferase AmbP1 apo structure | | Descriptor: | AmbP1, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z45
 
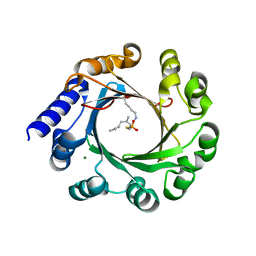 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5X18
 
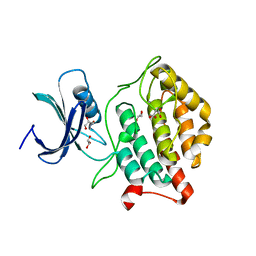 | | Crystal structure of Casein kinase I homolog 1 | | Descriptor: | Casein kinase I homolog 1, GLYCEROL, MALONIC ACID | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
5YY0
 
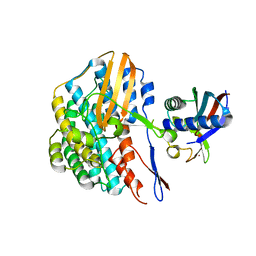 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YXY
 
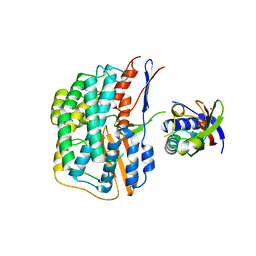 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Z44
 
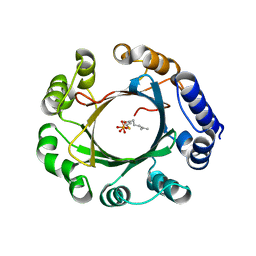 | | Crystal structure of prenyltransferase AmbP1 complexed with GSPP | | Descriptor: | AmbP1, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z46
 
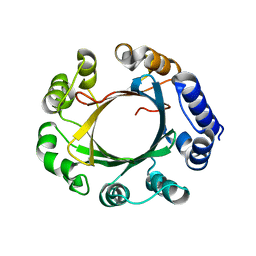 | | Crystal structure of prenyltransferase AmbP1 pH8 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2ZM0
 
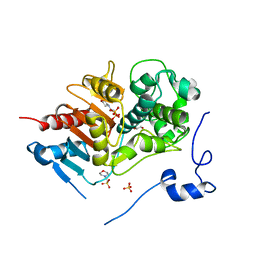 | | Structure of 6-aminohexanoate-dimer hydrolase, G181D/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
3VOU
 
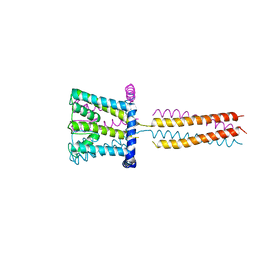 | | The crystal structure of NaK-NavSulP chimera channel | | Descriptor: | COBALT (II) ION, Ion transport 2 domain protein, Voltage-gated sodium channel, ... | | Authors: | Irie, K, Shimomura, T, Fujiyoshi, Y. | | Deposit date: | 2012-02-10 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate
Nat Commun, 3, 2012
|
|
3WVS
 
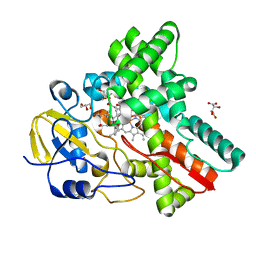 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
3WQP
 
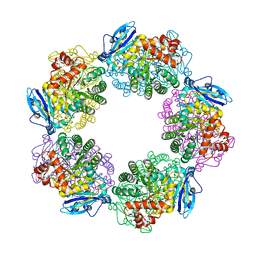 | | Crystal structure of Rubisco T289D mutant from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujihashi, M, Nishitani, Y, Kiriyama, T, Miki, K. | | Deposit date: | 2014-01-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mutation design of thermophilic Rubisco based on the three-dimensional structure enhances its activity at ambient temperature
to be published
|
|
3WKT
 
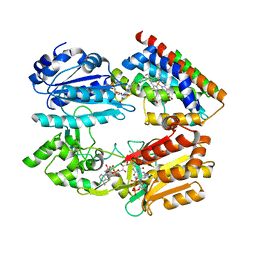 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZV2
 
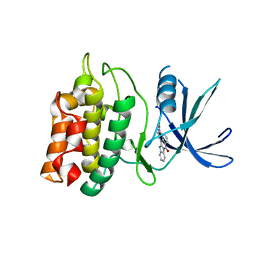 | | Crystal structure of human calcium/calmodulin-dependent protein kinase kinase 2, beta, CaMKK2 kinase domain in complex with STO-609 | | Descriptor: | 7-oxo-7H-benzimidazo[2,1-a]benz[de]isoquinoline-3-carboxylic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Yoshikawa, S, Kukimoto-niino, M, Shirouzu, M, Suzuki, A, Lee, S, Minokoshi, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-31 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Ca2+/calmodulin-dependent protein kinase kinase in complex with the inhibitor STO-609
J.Biol.Chem., 286, 2011
|
|
2CWN
 
 | | Crystal structure of mouse transaldolase | | Descriptor: | Transaldolase | | Authors: | Handa, N, Arai, R, Nishino, A, Uchikubo, T, Takemoto, C, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
