3AY7
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 G259A mutant | | Descriptor: | CHLORIDE ION, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
2ZQA
 
 | | Cefotaxime acyl-intermediate structure of class a beta-lacta Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | Beta-lactamase Toho-1, CEFOTAXIME, C3' cleaved, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
2ZQX
 
 | | Cytochrome P450BSbeta cocrystallized with heptanoic acid | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
2ZBS
 
 | |
2ZAS
 
 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with 4-alpha-cumylphenol, a bisphenol A derivative | | Descriptor: | 4-(1-methyl-1-phenylethyl)phenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ERRgamma tethers strongly bisphenol A and 4-alpha-cumylphenol in an induced-fit manner
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2E9M
 
 | | Crystal Structure of human Cytosolic Neutral beta-Glycosylceramidase (Klotho-related Prote:KLrP) complex with Galactose and fatty acids | | Descriptor: | Cytosolic beta-glucosidase, OLEIC ACID, PALMITIC ACID, ... | | Authors: | Kakuta, Y, Hayashi, Y, Okino, N, Ito, M. | | Deposit date: | 2007-01-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase.
J.Biol.Chem., 282, 2007
|
|
2ZQ9
 
 | | Cephalothin acyl-intermediate structure of class a beta-lactamase Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase Toho-1, CEPHALOTHIN, ... | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
2ZM2
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZK7
 
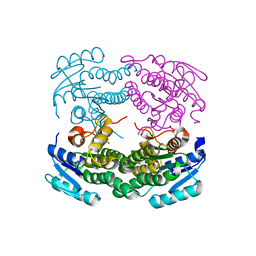 | | Structure of a C-terminal deletion mutant of Thermoplasma acidophilum aldohexose dehydrogenase (AldT) | | Descriptor: | Glucose 1-dehydrogenase related protein | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, N, Tamura, T. | | Deposit date: | 2008-03-12 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | C-terminal tail derived from the neighboring subunit is critical for the activity of Thermoplasma acidophilum D-aldohexose dehydrogenase
Proteins, 74, 2009
|
|
2Z5H
 
 | | Crystal structure of the head-to-tail junction of tropomyosin complexed with a fragment of TnT | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, Tropomyosin alpha-1 chain and General control protein GCN4, Troponin T, ... | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3A2F
 
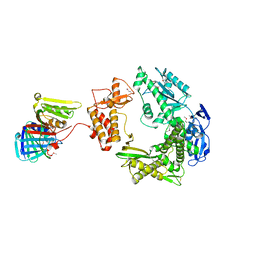 | |
3ABG
 
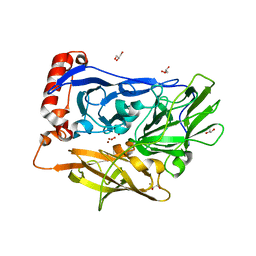 | | X-ray Crystal Analysis of Bilirubin Oxidase from Myrothecium verrucaria at 2.3 angstrom Resolution using a Twin Crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bilirubin oxidase, COPPER (II) ION, ... | | Authors: | Mizutani, K, Toyoda, M, Sagara, K, Takahashi, N, Sato, A, Kamitaka, Y, Tsujimura, S, Nakanishi, Y, Sugiura, T, Yamaguchi, S, Kano, K, Mikami, B. | | Deposit date: | 2009-12-10 | | Release date: | 2010-08-18 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of bilirubin oxidase from Myrothecium verrucaria at 2.3 A resolution using a twinned crystal
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3AUS
 
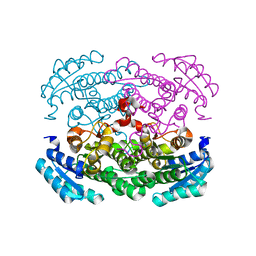 | |
3APN
 
 | | Crystal structure of the human wild-type PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3AY6
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 A258F mutant in complex with NADH and D-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Glucose 1-dehydrogenase 4, ... | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-04-29 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AUT
 
 | | Crystal structure of Bacillus megaterium glucose dehydrogenase 4 in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glucose 1-dehydrogenase 4 | | Authors: | Nishioka, T, Yasutake, Y, Nishiya, Y, Tamura, T. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided mutagenesis for the improvement of substrate specificity of Bacillus megaterium glucose 1-dehydrogenase IV
Febs J., 279, 2012
|
|
3AWM
 
 | | Cytochrome P450SP alpha (CYP152B1) wild-type with palmitic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position
J.Biol.Chem., 286, 2011
|
|
3AWP
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant F288G | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
3AWQ
 
 | | Cytochrome P450SP alpha (CYP152B1) mutant L78F | | Descriptor: | Fatty acid alpha-hydroxylase, PALMITIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position.
J.Biol.Chem., 286, 2011
|
|
2E9L
 
 | | Crystal Structure of human Cytosolic Neutral beta-Glycosylceramidase (Klotho-related Prote:KLrP) complex with Glucose and fatty acids | | Descriptor: | Cytosolic beta-glucosidase, GLYCEROL, OLEIC ACID, ... | | Authors: | Kakuta, Y, Hayashi, Y, Okino, N, Ito, M. | | Deposit date: | 2007-01-25 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase.
J.Biol.Chem., 282, 2007
|
|
3AGF
 
 | |
3A16
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Propionaldoxime | | Descriptor: | (1Z)-propanal oxime, Aldoxime dehydratase, MAGNESIUM ION, ... | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A18
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (soaked crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A17
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (Co-crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A15
 
 | | Crystal Structure of Substrate-Free Form of Aldoxime Dehydratase (OxdRE) | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
