6YYW
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | 2-OXOGLUTARIC ACID, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6YYY
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 4,4-dimethyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | 2,2-dimethyl-4-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6Z6Q
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 3-ethyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | (3~{R})-3-ethyl-2-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6YYV
 
 | |
6Z6R
 
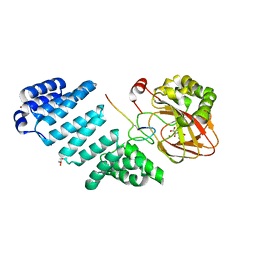 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, N-oxalyl-alpha-methylalanine, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, BROMIDE ION, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6YYU
 
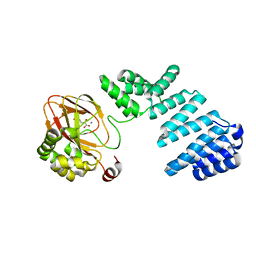 | |
6YYX
 
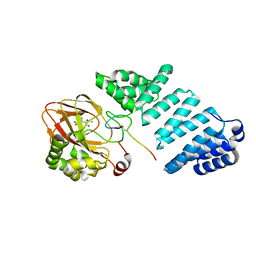 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 3-methyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | (3~{R})-3-methyl-2-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase
Chem Sci, 12, 2021
|
|
4XD7
 
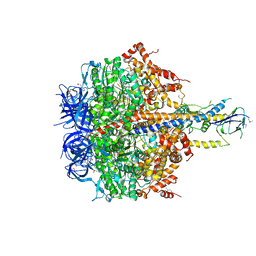 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
1YGS
 
 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
7R8J
 
 | |
8WD4
 
 | | EGFR(L858R/T790/C797S) in complex with compound 5j | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, ~{N}-[3,3-bis(fluoranyl)propyl]-4-[[(2~{S})-butan-2-yl]amino]-6-[[2-(1-cyclopropylsulfonylpyrazol-4-yl)pyrimidin-4-yl]amino]pyridine-3-carboxamide | | Authors: | Nishikawa, Y. | | Deposit date: | 2023-09-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis, activity, and their relationships of 2,4-diaminonicotinamide derivatives as EGFR inhibitors targeting C797S mutation.
Bioorg.Med.Chem.Lett., 98, 2023
|
|
6WOX
 
 | |
6WOY
 
 | |
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
2RSE
 
 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
6RUJ
 
 | |
6T1H
 
 | | OXA-51-like beta-lactamase OXA-66 | | Descriptor: | Beta-lactamase OXA-66, ZINC ION | | Authors: | Takebayashi, Y, Chirgadze, D, Henderson, S, Warburton, P.J, Evans, B.E. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the OXA-51-like beta-lactamase OXA-66
To Be Published
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
4YN5
 
 | | Catalytic domain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase | | Descriptor: | CACODYLATE ION, Mannan endo-1,4-beta-mannosidase | | Authors: | Shimane, Y, Ohta, Y, Usami, R, Hatada, Y. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase
To Be Published
|
|
6OY7
 
 | |
6OVY
 
 | |
6OVR
 
 | |
6OW3
 
 | |
6OY6
 
 | |
6OY5
 
 | |
