1WG8
 
 | |
1H6L
 
 | |
1GS3
 
 | |
8H4V
 
 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
8HB5
 
 | | Crystal structure of Mincle in complex with HD-275 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
1F07
 
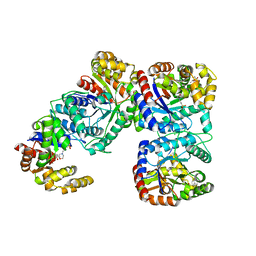 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1EZW
 
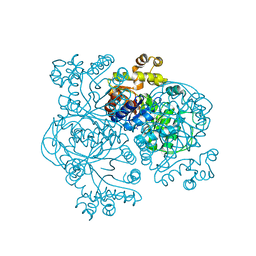 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
3RK8
 
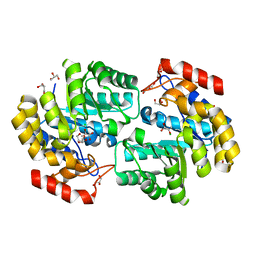 | | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kaushik, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-17 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution
To be Published
|
|
2RRF
 
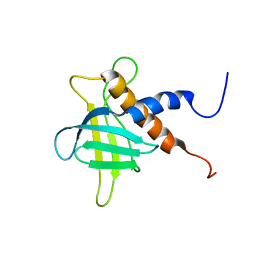 | | The solution structure of the C-terminal region of Zinc finger FYVE domain-containing protein 21 | | Descriptor: | Zinc finger FYVE domain-containing protein 21 | | Authors: | Koshiba, S, Tomizawa, T, Hayashi, F, Tochio, N, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-08-03 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ZF21 protein, a regulator of the disassembly of focal adhesions and cancer metastasis, contains a novel noncanonical pleckstrin homology domain
J.Biol.Chem., 286, 2011
|
|
5WVO
 
 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
6ICS
 
 | |
6IDC
 
 | |
6IF6
 
 | | Structure of the periplasmic domain of SflA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein SflA | | Authors: | Nishikawa, S, Sakuma, M, Kojima, S, Homma, M, Imada, K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the periplasmic domain of SflA involved in spatial regulation of the flagellar biogenesis of Vibrio reveals a TPR/SLR-like fold.
J.Biochem., 166, 2019
|
|
6IYS
 
 | |
4JWK
 
 | | Crystal structure of the complex of peptidyl-tRNA hydrolase from Acinetobacter baumannii with cytidine at 1.87 A resolution | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
6IEI
 
 | |
4JX9
 
 | | Crystal structure of the complex of peptidyl t-RNA hydrolase from Acinetobacter baumannii with uridine at 1.4A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, URIDINE | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-28 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
4LWQ
 
 | | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Kaushik, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-28 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native peptidyl t-RNA hydrolase from Acinetobacter baumannii at 1.38A resolution
To be Published
|
|
2RVA
 
 | |
2RV9
 
 | |
2ECC
 
 | | Solution Structure of the second Homeobox Domain of Human Homeodomain Leucine Zipper-Encoding Gene (Homez) | | Descriptor: | Homeobox and leucine zipper protein Homez | | Authors: | Ohnishi, S, Kamatari, Y.O, Tochio, N, Nameki, N, Miyamoto, K, Li, H, Kobayashi, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-13 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the second Homeobox Domain of Human Homeodomain Leucine Zipper-Encoding Gene (Homez)
To be Published
|
|
8JT1
 
 | | COLLAGENASE FROM GRIMONTIA (VIBRIO) HOLLISAE 1706B COMPLEXED WITH GLY-PRO-HYP-GLY-PRO-HYP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-mer peptide, ... | | Authors: | Ueshima, S, Yaskawa, K, Takita, T, Mikami, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the catalytic mechanism of Grimontia hollisae collagenase through structural and mutational analyses.
Febs Lett., 597, 2023
|
|
3QRC
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis, in complex with the heparin analogue sucrose octasulfate | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
3QRA
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
5XE9
 
 | | Crystal Structure of the Complex of the Peptidase Domain of Streptococcus mutans ComA with a Small Molecule Inhibitor. | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA, [(1~{S},2~{R},4~{S},5~{R})-5-[5-(4-methoxyphenyl)-2-methyl-pyrazol-3-yl]-1-azabicyclo[2.2.2]octan-2-yl]methyl ~{N}-propylcarbamate | | Authors: | Ishii, S, Fukui, K, Yokoshima, S, Kumagai, K, Beniyama, Y, Kodama, T, Fukuyama, T, Okabe, T, Nagano, T, Kojima, H, Yano, T. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | High-throughput Screening of Small Molecule Inhibitors of the Streptococcus Quorum-sensing Signal Pathway
Sci Rep, 7, 2017
|
|
