2D8C
 
 | | Solution structure of the sam-domain of mouse phosphatidyl ceramidecholinephosphotransferase 1 | | 分子名称: | Phosphatidylcholine:ceramide cholinephosphotransferase 1 | | 著者 | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-12-02 | | 公開日 | 2006-06-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the sam-domain of mouse phosphatidyl ceramidecholinephosphotransferase 1
To be Published
|
|
7EXW
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with alpha-L-arabinofuranosylamide | | 分子名称: | 2-bromanyl-N-[(2R,3R,4R,5S}-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXU
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 E322Q mutant complexed with p-nitrophenyl beta-L-arabinofuranoside | | 分子名称: | (2S,3R,4R,5R)-2-(hydroxymethyl)-5-(4-nitrophenoxy)oxolane-3,4-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Maruyama, S, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
7EXV
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranoylamide | | 分子名称: | 2-bromanyl-N-[(2S,3R,4R,5S)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]ethanamide, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | 著者 | Sawano, K, Arakawa, T, Yamada, C, Fujita, K, Fushinobu, S. | | 登録日 | 2021-05-28 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase.
Glycobiology, 32, 2022
|
|
3K2U
 
 | | Crystal structure of HGFA in complex with the allosteric inhibitory antibody Fab40 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Fab fragment, ... | | 著者 | Ganesan, R, Eigenbrot, C, Shia, S. | | 登録日 | 2009-09-30 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Unraveling the allosteric mechanism of serine protease inhibition by an antibody.
Structure, 17, 2009
|
|
5ZYH
 
 | | Crystal structure of CERT START domain in complex with compound E5 | | 分子名称: | 2-[4-[3-~{tert}-butyl-5-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYK
 
 | | Crystal structure of CERT START domain in complex with compound E25 | | 分子名称: | 2-[4-[4-cyclopentyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5ZYG
 
 | | Crystal structure of CERT START domain in complex with compound B5 | | 分子名称: | 2-[4-[3-~{tert}-butyl-5-(2-pyridin-2-ylethyl)phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
1WQ3
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with 3-iodo-L-tyrosine | | 分子名称: | 3-IODO-TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5ZYM
 
 | | Crystal structure of CERT START domain in complex with compound E25B | | 分子名称: | 2-[4-[4-cyclopentyl-3-[(1~{R},2~{S})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, GLYCEROL, LIPID-TRANSFER PROTEIN CERT | | 著者 | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | 登録日 | 2018-05-25 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
1QB4
 
 | | CRYSTAL STRUCTURE OF MN(2+)-BOUND PHOSPHOENOLPYRUVATE CARBOXYLASE | | 分子名称: | ASPARTIC ACID, MANGANESE (II) ION, PHOSPHOENOLPYRUVATE CARBOXYLASE | | 著者 | Matsumura, H, Terada, M, Shirakata, S, Inoue, T, Yoshinaga, T, Izui, K, Kai, Y. | | 登録日 | 1999-04-30 | | 公開日 | 2002-05-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Plausible phosphoenolpyruvate binding site revealed by 2.6 A structure of Mn2+-bound phosphoenolpyruvate carboxylase from Escherichia coli
FEBS Lett., 458, 1999
|
|
1WQ4
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with L-tyrosine | | 分子名称: | TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6HF6
 
 | | Crystal structure of the Protease 1 (E29A,E60A,E80A) from Pyrococcus horikoshii co-crystallized with Tb-Xo4. | | 分子名称: | Deglycase PH1704, MALONATE ION, TERBIUM(III) ION, ... | | 著者 | Engilberge, S, Wagner, T, Santoni, G, Breyton, C, Shima, S, Franzetti, B, Riobe, F, Maury, O, Girard, E. | | 登録日 | 2018-08-21 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Protein crystal structure determination with the crystallophore, a nucleating and phasing agent.
J.Appl.Crystallogr., 52, 2019
|
|
3VJM
 
 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | 分子名称: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | 登録日 | 2011-10-24 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
8H0I
 
 | | Cryo-EM structure of APOBEC3G-Vif complex | | 分子名称: | APOBEC3G, CHLORIDE ION, Core binding factor beta, ... | | 著者 | Kouno, T, Shibata, S, Hyun, J, Kim, T.G, Wolf, M. | | 登録日 | 2022-09-29 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into RNA bridging between HIV-1 Vif and antiviral factor APOBEC3G.
Nat Commun, 14, 2023
|
|
1X8X
 
 | | Tyrosyl t-RNA Synthetase from E.coli Complexed with Tyrosine | | 分子名称: | SULFATE ION, TYROSINE, Tyrosyl-tRNA synthetase | | 著者 | Kobayashi, T, Takimura, T, Sekine, R, Kelly, V.P, Kamata, K, Sakamoto, K, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-08-19 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Snapshots of the KMSKS Loop Rearrangement for Amino Acid Activation by Bacterial Tyrosyl-tRNA Synthetase
J.MOL.BIOL., 346, 2005
|
|
7E5V
 
 | | Crystal structure of Phm7 in complex with inhibitor | | 分子名称: | Diels-Alderase, GLYCEROL, SULFATE ION, ... | | 著者 | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | 登録日 | 2021-02-20 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5T
 
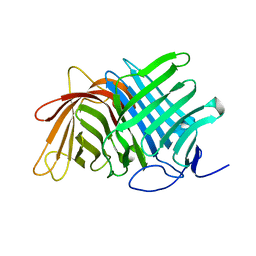 | | Crystal structure of Fsa2 | | 分子名称: | Diels-Alderase fsa2, ETHANOL, PENTAETHYLENE GLYCOL, ... | | 著者 | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | 登録日 | 2021-02-20 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.16977525 Å) | | 主引用文献 | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5U
 
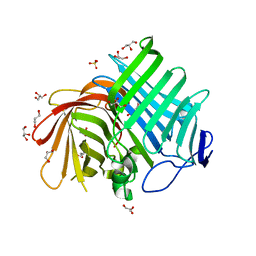 | | Crystal structure of Phm7 | | 分子名称: | CHLORIDE ION, Diels-Alderase, GLYCEROL, ... | | 著者 | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | 登録日 | 2021-02-20 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8HEW
 
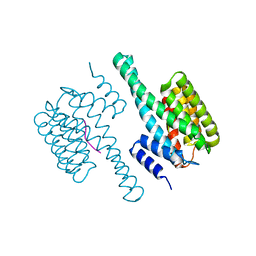 | | Potato 14-3-3 St14f | | 分子名称: | 14-3-3 protein, StFDL1 peptide | | 著者 | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | 登録日 | 2022-11-08 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
5EF9
 
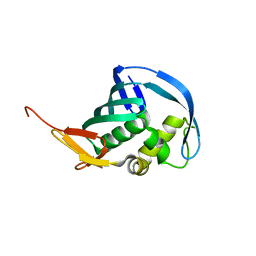 | |
4JAW
 
 | | Crystal Structure of Lacto-N-Biosidase from Bifidobacterium bifidum complexed with LNB-thiazoline | | 分子名称: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Lacto-N-biosidase, SULFATE ION, ... | | 著者 | Ito, T, Katayama, T, Stubbs, K.A, Fushinobu, S. | | 登録日 | 2013-02-19 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of a glycoside hydrolase family 20 lacto-N-biosidase from Bifidobacterium bifidum
J.Biol.Chem., 288, 2013
|
|
5EFA
 
 | |
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | 登録日 | 2023-01-19 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
6HK1
 
 | | Crystal structure of the Thiazole synthase from Methanothermococcus thermolithotrophicus co-crystallized with Tb-Xo4 | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SODIUM ION, ... | | 著者 | Engilberge, S, Wagner, T, Santoni, G, Breyton, C, Shima, S, Franzetti, B, Riobe, F, Maury, O, Girard, E. | | 登録日 | 2018-09-05 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Protein crystal structure determination with the crystallophore, a nucleating and phasing agent.
J.Appl.Crystallogr., 52, 2019
|
|
