4ZLF
 
 | | Cellobionic acid phosphorylase - cellobionic acid complex | | 分子名称: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4YHJ
 
 | | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4 (GRK4) | | 分子名称: | AMP PHOSPHORAMIDATE, G protein-coupled receptor kinase 4 | | 著者 | Allen, S.J, Parthasarathy, G, Soisson, S, Munshi, S. | | 登録日 | 2015-02-27 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4.
J.Biol.Chem., 290, 2015
|
|
7XMC
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | 登録日 | 2022-04-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMD
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4 | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | 登録日 | 2022-04-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
5H41
 
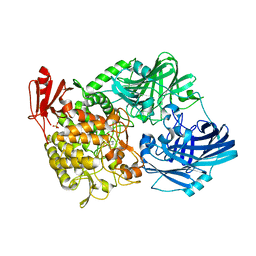 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | 分子名称: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
5H40
 
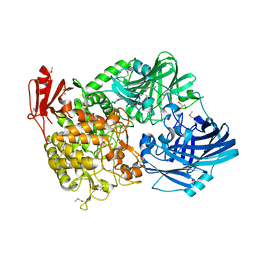 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose | | 分子名称: | CALCIUM ION, GLYCEROL, Uncharacterized protein, ... | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
1JAY
 
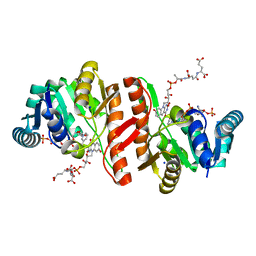 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) with its substrates bound | | 分子名称: | COENZYME F420, Coenzyme F420H2:NADP+ Oxidoreductase (FNO), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | 登録日 | 2001-06-01 | | 公開日 | 2001-12-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
4ZLI
 
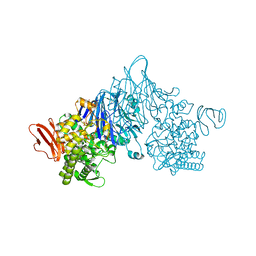 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
2Z6W
 
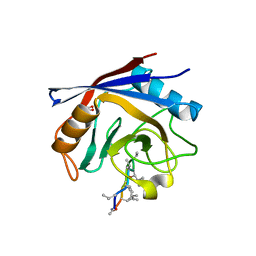 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | 分子名称: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | 著者 | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | 登録日 | 2007-08-09 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.96 Å) | | 主引用文献 | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
4ZLE
 
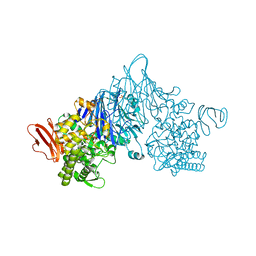 | | Cellobionic acid phosphorylase - ligand free structure | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | 分子名称: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | 著者 | Ito, T, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-06-09 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
6IMF
 
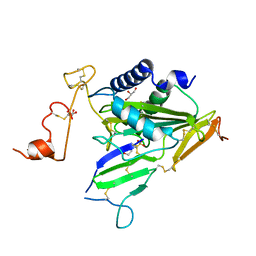 | | Crystal structure of TOXIN/ANTITOXIN complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cysteine-rich venom protein triflin, GLYCEROL, ... | | 著者 | Shioi, N, Tadokoro, T, Shioi, S, Hu, Y, Kurahara, L.H, Okabe, Y, Matsubara, H, Kita, S, Ose, T, Kuroki, K, Maenaka, K, Terada, S. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the complex between venom toxin and serum inhibitor from Viperidae snake.
J. Biol. Chem., 294, 2019
|
|
1MG8
 
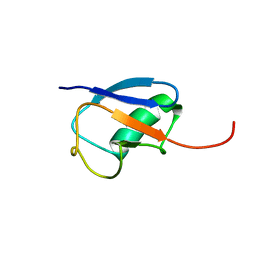 | | NMR structure of ubiquitin-like domain in murine Parkin | | 分子名称: | Parkin | | 著者 | Tashiro, M, Okubo, S, Shimotakahara, S, Hatanaka, H, Yasuda, H, Kainosho, M, Yokoyama, S, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-08-15 | | 公開日 | 2003-04-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of ubiquitin-like domain in PARKIN: Gene product of familial Parkinson's disease.
J.Biomol.NMR, 25, 2003
|
|
1KFA
 
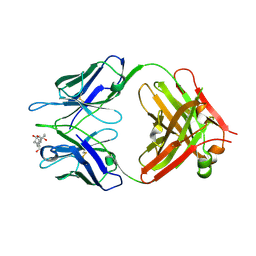 | | Crystal structure of Fab fragment complexed with gibberellin A4 | | 分子名称: | GIBBERELLIN A4, monoclonal antibody heavy chain, monoclonal antibody light chain | | 著者 | Murata, T, Fushinobu, S, Nakajima, M, Asami, O, Sassa, T, Wakagi, T, Yamaguchi, I. | | 登録日 | 2001-11-20 | | 公開日 | 2002-09-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the liganded anti-gibberellin A(4) antibody 4-B8(8)/E9 Fab fragment.
Biochem.Biophys.Res.Commun., 293, 2002
|
|
1UG7
 
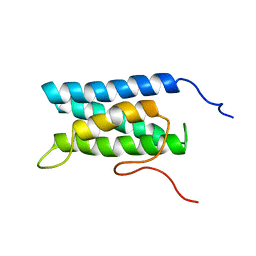 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | 分子名称: | 2610208M17Rik protein | | 著者 | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-06-13 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
8XE7
 
 | |
8WM0
 
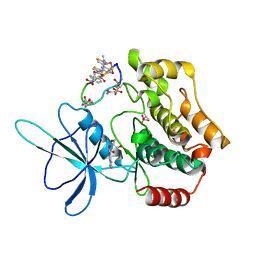 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | 分子名称: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | 著者 | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2023-10-01 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
4BS9
 
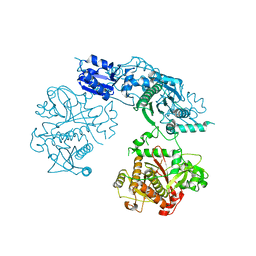 | | Structure of the heterocyclase TruD | | 分子名称: | TRUD, ZINC ION | | 著者 | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | 登録日 | 2013-06-09 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1BQR
 
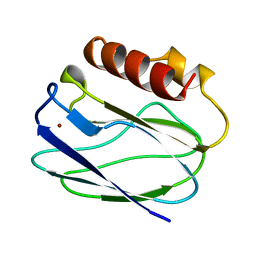 | | REDUCED PSEUDOAZURIN | | 分子名称: | COPPER (II) ION, PSEUDOAZURIN | | 著者 | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | 登録日 | 1998-08-17 | | 公開日 | 1999-08-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure determinations of oxidized and reduced pseudoazurins from Achromobacter cycloclastes. Concerted movement of copper site in redox forms with the rearrangement of hydrogen bond at a remote histidine.
J.Biol.Chem., 274, 1999
|
|
1X67
 
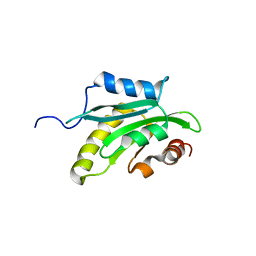 | | Solution structure of the cofilin homology domain of HIP-55 (drebrin-like protein) | | 分子名称: | Drebrin-like protein | | 著者 | Goroncy, A.K, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-17 | | 公開日 | 2005-11-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
3E5J
 
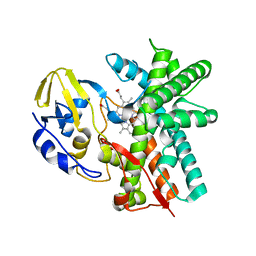 | | Crystal structure of CYP105P1 wild-type ligand-free form | | 分子名称: | Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | 登録日 | 2008-08-14 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
4AKS
 
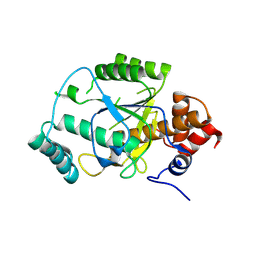 | | PatG macrocyclase domain | | 分子名称: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | 著者 | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | 登録日 | 2012-02-28 | | 公開日 | 2012-07-18 | | 最終更新日 | 2013-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6KNC
 
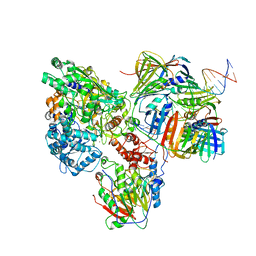 | | PolD-PCNA-DNA (form B) | | 分子名称: | DNA polymerase D DP2 (DNA polymerase II large) subunit, DNA polymerase II small subunit, DNA polymerase sliding clamp 1, ... | | 著者 | Mayanagi, K, Oki, K, Miyazaki, N, Ishino, S, Yamagami, T, Iwasaki, K, Kohda, D, Morikawa, K, Shirai, T, Ishino, Y. | | 登録日 | 2019-08-05 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (9.3 Å) | | 主引用文献 | Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy.
Bmc Biol., 18, 2020
|
|
7CP6
 
 | | Crystal structure of FqzB | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | 著者 | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | 登録日 | 2020-08-06 | | 公開日 | 2020-12-30 | | 最終更新日 | 2021-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
1BQT
 
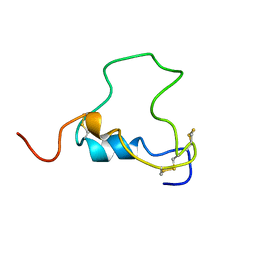 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | 分子名称: | INSULIN-LIKE GROWTH FACTOR-I | | 著者 | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | 登録日 | 1998-08-18 | | 公開日 | 1999-05-18 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
