5IB0
 
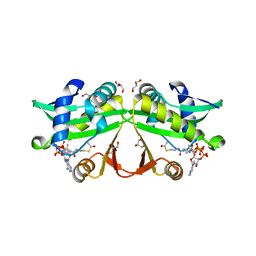 | | PA4534: acetyl CoA complex | | 分子名称: | ACETYL COENZYME *A, GLYCEROL, Uncharacterized protein PA4534 | | 著者 | Choe, J, Shin, S. | | 登録日 | 2016-02-22 | | 公開日 | 2017-03-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of PA4534
To Be Published
|
|
7YQS
 
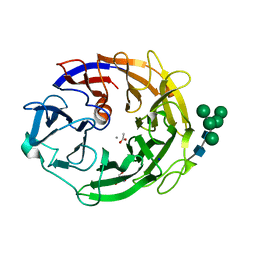 | | Neutron structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | 登録日 | 2022-08-08 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | NEUTRON DIFFRACTION (1.25 Å), X-RAY DIFFRACTION | | 主引用文献 | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
3F9N
 
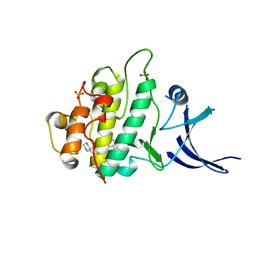 | | Crystal structure of chk1 kinase in complex with inhibitor 38 | | 分子名称: | 3-(3-chlorophenyl)-2-({(1S)-1-[(6S)-2,8-diazaspiro[5.5]undec-2-ylcarbonyl]pentyl}sulfanyl)quinazolin-4(3H)-one, SULFATE ION, Serine/threonine-protein kinase Chk1 | | 著者 | Yan, Y, Munshi, S, Ikuta, M. | | 登録日 | 2008-11-14 | | 公開日 | 2009-01-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development of thioquinazolinones, allosteric Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4YRD
 
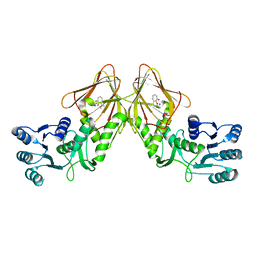 | | Crystal structure of CapF with inhibitor 3-isopropenyl-tropolone | | 分子名称: | 2-hydroxy-3-(prop-1-en-2-yl)cyclohepta-2,4,6-trien-1-one, Capsular polysaccharide synthesis enzyme Cap5F, ZINC ION | | 著者 | Nakano, K, Chigira, T, Miyafusa, T, Nagatoishi, S, Caaveiro, J.M.M, Tsumoto, K. | | 登録日 | 2015-03-15 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Discovery and characterization of natural tropolones as inhibitors of the antibacterial target CapF from Staphylococcus aureus.
Sci Rep, 5, 2015
|
|
5CI9
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 6 | | 分子名称: | 1-(propan-2-yl)-1H-benzimidazole-5-carboxylic acid, Protein Tob1, SODIUM ION | | 著者 | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | 登録日 | 2015-07-11 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
4YHJ
 
 | | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4 (GRK4) | | 分子名称: | AMP PHOSPHORAMIDATE, G protein-coupled receptor kinase 4 | | 著者 | Allen, S.J, Parthasarathy, G, Soisson, S, Munshi, S. | | 登録日 | 2015-02-27 | | 公開日 | 2015-07-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure and Function of the Hypertension Variant A486V of G Protein-coupled Receptor Kinase 4.
J.Biol.Chem., 290, 2015
|
|
5AVF
 
 | | The ligand binding domain of Mlp37 with taurine | | 分子名称: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | 著者 | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2015-06-15 | | 公開日 | 2016-06-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
6JT3
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Tadano, G, Komano, K, Yoshida, S, Suzuki, S, Nakahara, K, Fuchino, K, Fujimoto, K, Matsuoka, E, Yamamoto, T, Asada, N, Ito, H, Sakaguchi, G, Kanegawa, N, Kido, Y, Ando, S, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Bergh, A.V.D, Austin, N, Gijsen, H.J.M, Yamano, Y, Iso, Y, Kusakabe, K.I. | | 登録日 | 2019-04-08 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of an Extremely Potent Thiazine-Based beta-Secretase Inhibitor with Reduced Cardiovascular and Liver Toxicity at a Low Projected Human Dose.
J.Med.Chem., 62, 2019
|
|
3CZA
 
 | | Crystal Structure of E18D DJ-1 | | 分子名称: | MALONIC ACID, Protein DJ-1 | | 著者 | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hashim, S, Pozharski, E, Wilson, M.A. | | 登録日 | 2008-04-28 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
5CI8
 
 | | Crystal structure of human Tob in complex with inhibitor fragment 1 | | 分子名称: | Protein Tob1, pyrrolo[1,2-a]quinoxalin-4(5H)-one | | 著者 | Bai, Y, Tashiro, S, Nagatoishi, S, Suzuki, T, Tsumoto, K, Bartlam, M, Yamamoto, T. | | 登録日 | 2015-07-11 | | 公開日 | 2015-11-18 | | 最終更新日 | 2015-12-09 | | 実験手法 | X-RAY DIFFRACTION (2.328 Å) | | 主引用文献 | Structural basis for inhibition of the Tob-CNOT7 interaction by a fragment screening approach
Protein Cell, 6, 2015
|
|
3N4N
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3' | | 著者 | Takenaka, A, Juan, E.C.M, Shimizu, S. | | 登録日 | 2010-05-22 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
8JQP
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with 3,4-dihydroxybenzoate | | 分子名称: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, ... | | 著者 | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | 登録日 | 2023-06-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQO
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus complexed with imidazole | | 分子名称: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | 登録日 | 2023-06-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
8JQQ
 
 | | Protocatecuate hydroxylase from Xylophilus ampelinus C347T mutant | | 分子名称: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Fukushima, R, Katsuki, N, Fushinobu, S, Takaya, N. | | 登録日 | 2023-06-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Protocatechuate hydroxylase is a novel group A flavoprotein monooxygenase with a unique substrate recognition mechanism.
J.Biol.Chem., 300, 2023
|
|
3N4O
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | 分子名称: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*(B7C)P*CP*GP*CP*G)-3' | | 著者 | Takenaka, A, Juan, E.C.M, Shimizu, S. | | 登録日 | 2010-05-22 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
1V61
 
 | | Solution Structure of the Pleckstrin Homology Domain of alpha-Pix | | 分子名称: | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | | 著者 | Nakanishi, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-26 | | 公開日 | 2004-05-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Pleckstrin Homology Domain of alpha-Pix
To be Published
|
|
6IMF
 
 | | Crystal structure of TOXIN/ANTITOXIN complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cysteine-rich venom protein triflin, GLYCEROL, ... | | 著者 | Shioi, N, Tadokoro, T, Shioi, S, Hu, Y, Kurahara, L.H, Okabe, Y, Matsubara, H, Kita, S, Ose, T, Kuroki, K, Maenaka, K, Terada, S. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the complex between venom toxin and serum inhibitor from Viperidae snake.
J. Biol. Chem., 294, 2019
|
|
4ZLF
 
 | | Cellobionic acid phosphorylase - cellobionic acid complex | | 分子名称: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Abe, K, Yokoshima, S, Yoshimori, A. | | 登録日 | 2023-02-28 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.08 Å) | | 主引用文献 | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJW
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-06 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Abe, K, Yokoshima, S, Yoshimori, A. | | 登録日 | 2023-02-28 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.19 Å) | | 主引用文献 | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJV
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | 著者 | Abe, K, Yokoshima, S, Yoshimori, A. | | 登録日 | 2023-02-28 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8JMN
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-21 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[[2-[(4-chlorophenyl)methoxy]phenyl]methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Abe, K, Yokoshima, S, Yoshimori, A. | | 登録日 | 2023-06-05 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | 分子名称: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | 著者 | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | 登録日 | 2023-11-19 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V2N
 
 | | Human SIRT3 co-crystallized with ligands, including p53-AMC peptide and Carba-NAD | | 分子名称: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, NAD-dependent protein deacetylase sirtuin-3, ... | | 著者 | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | 登録日 | 2023-11-23 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
4ZLI
 
 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
