1OMG
 
 | | NMR STUDY OF OMEGA-CONOTOXIN MVIIA | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Kohno, T, Kim, J.-I, Kobayashi, K, Kodera, Y, Maeda, T, Sato, K. | | Deposit date: | 1995-04-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the calcium channel blocker omega-conotoxin MVIIA.
Biochemistry, 34, 1995
|
|
7WAG
 
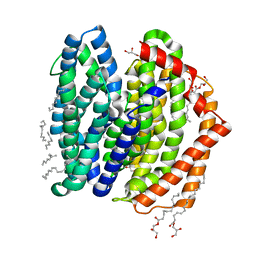 | | Crystal structure of MurJ squeezed form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAW
 
 | | MurJ inward closed form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAX
 
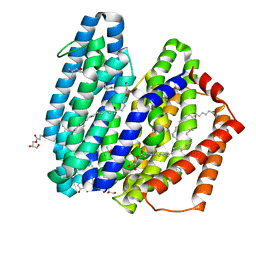 | | MurJ inward occluded form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, lipid II flippase MurJ | | Authors: | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
5IP3
 
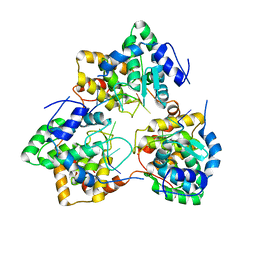 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssDNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
1FUT
 
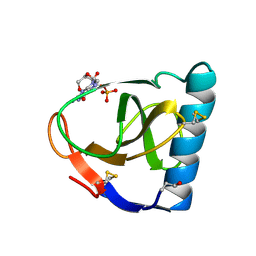 | | CRYSTAL STRUCTURES OF RIBONUCLEASE F1 OF FUSARIUM MONILIFORME IN ITS FREE FORM AND IN COMPLEX WITH 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE F1 | | Authors: | Katayanagi, K, Vassylyev, D.G, Ishikawa, K, Morikawa, K. | | Deposit date: | 1993-01-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of ribonuclease F1 of Fusarium moniliforme in its free form and in complex with 2'GMP.
J.Mol.Biol., 230, 1993
|
|
5IP2
 
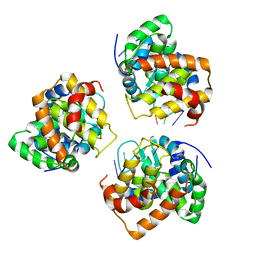 | | Tomato spotted wilt tospovirus nucleocapsid protein-ssRNA complex | | Descriptor: | Nucleoprotein, RNA (5'-D(P*UP*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
1G0D
 
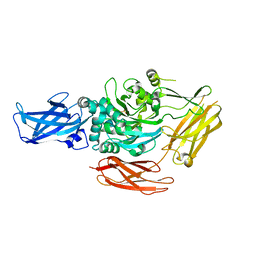 | | CRYSTAL STRUCTURE OF RED SEA BREAM TRANSGLUTAMINASE | | Descriptor: | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, SULFATE ION | | Authors: | Noguchi, K, Ishikawa, K, Yokoyama, K, Ohtsuka, T, Nio, N, Suzuki, E. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of red sea bream transglutaminase.
J.Biol.Chem., 276, 2001
|
|
6D6S
 
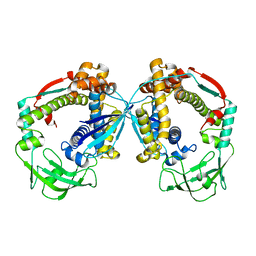 | |
2K4H
 
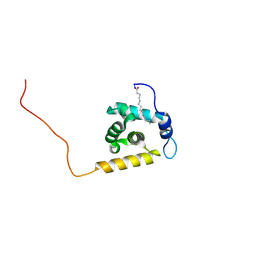 | | Solution structure of the HIV-2 myristoylated Matrix protein | | Descriptor: | HIV-2 myristoylated matrix protein, MYRISTIC ACID | | Authors: | Saad, J.S, Ablan, S.D, Ghanam, R.H, Kim, A, Andrews, K, Nagashima, K, Freed, E.O, Summers, M.F. | | Deposit date: | 2008-06-08 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting.
J.Mol.Biol., 382, 2008
|
|
1FUS
 
 | |
2JMM
 
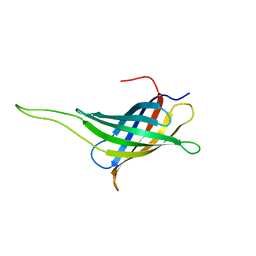 | | NMR solution structure of a minimal transmembrane beta-barrel platform protein | | Descriptor: | Outer membrane protein A | | Authors: | Johansson, M.U, Alioth, S, Hu, K, Walser, R, Koebnik, R, Pervushin, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance
Biochemistry, 46, 2007
|
|
2K4I
 
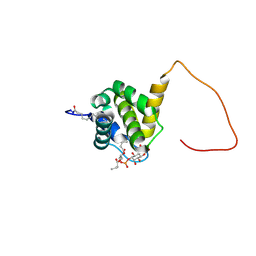 | | Solution structure of HIV-2 myrMA bound to di-C4-PI(4,5)P2 | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1,2-DIYL DIBUTANOATE, HIV-2 myristoylated matrix protein, MYRISTIC ACID | | Authors: | Saad, J.S, Ablan, S.D, Ghanam, R.H, Kim, A, Andrews, K, Nagashima, K, Freed, E.O, Summers, M.F. | | Deposit date: | 2008-06-08 | | Release date: | 2008-08-12 | | Last modified: | 2014-01-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting.
J.Mol.Biol., 382, 2008
|
|
5IP1
 
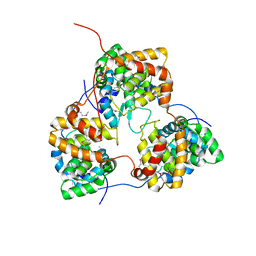 | | Tomato spotted wilt tospovirus nucleocapsid protein | | Descriptor: | Nucleoprotein | | Authors: | Komoda, K, Narita, M, Yamashita, K, Tanaka, I, Yao, M. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus.
J. Virol., 91, 2017
|
|
2K4E
 
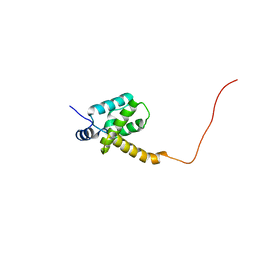 | | Solution structure of the HIV-2 UNMYRISTOYLATED MATRIX PROTEIN | | Descriptor: | HIV-2 unmyristoylated matrix protein | | Authors: | Saad, J.S, Ablan, S.D, Ghanam, R.H, Kim, A, Andrews, K, Nagashima, K, Freed, E.O, Summers, M.F. | | Deposit date: | 2008-06-07 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting.
J.Mol.Biol., 382, 2008
|
|
4HE7
 
 | | Crystal Structure of Brazzein | | Descriptor: | Defensin-like protein, SODIUM ION | | Authors: | Nagata, K, Hongo, N, Kameda, Y, Yamamura, A, Sasaki, H, Lee, W.C, Ishikawa, K, Suzuki, E, Tanokura, M. | | Deposit date: | 2012-10-03 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5YHA
 
 | | Crystal structure of Pd(allyl)/Wild Type Polyhedra | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PALLADIUM ION, ... | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
2LHI
 
 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
1IQA
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MOUSE RANK LIGAND | | Descriptor: | RECEPTOR ACTIVATOR OF NUCLEAR FACTOR KAPPA B LIGAND | | Authors: | Ito, S, Wakabayashi, K, Ubukata, O, Hayashi, S, Okada, F, Hata, T. | | Deposit date: | 2001-07-11 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the extracellular domain of mouse RANK ligand at 2.2-A resolution.
J.Biol.Chem., 277, 2002
|
|
8HGA
 
 | |
8HIA
 
 | |
5YHB
 
 | | Crystal structure of Pd(allyl)/polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | PALLADIUM ION, Palladium(II) allyl complex, Polyhedrin | | Authors: | Abe, S, Atsumi, K, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of in cell protein crystals containing organometallic complexes.
Phys Chem Chem Phys, 20, 2018
|
|
1J03
 
 | | Solution structure of a putative steroid-binding protein from Arabidopsis | | Descriptor: | putative steroid binding protein | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Terada, T, Shirouzu, M, Seki, M, Shinozaki, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-29 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an Arabidopsis homologue of the mammalian membrane-associated progesterone receptor
To be Published
|
|
6JU6
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant | | Descriptor: | NITRATE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
