7UU0
 
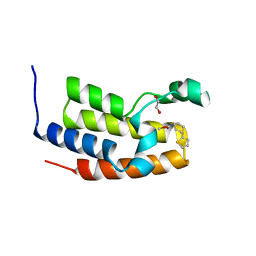 | | Crystal structure of the BRD2-BD2 in complex with a ligand | | Descriptor: | 1,2-ETHANEDIOL, Isoform 3 of Bromodomain-containing protein 2, methyl (7S)-7-(thiophen-2-yl)-1,4-thiazepane-4-carboxylate | | Authors: | Kalra, P, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the BRD2-BD2 in complex with a ligand
To Be Published
|
|
8UZF
 
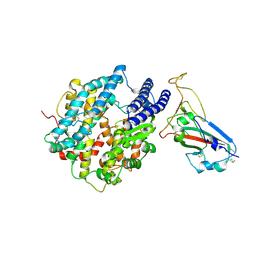 | | Crystal structure of chimeric RaTG13 RBD complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Crystal structure of chimeric RaTG13 RBD complexed with chimeric mouse ACE2
To Be Published
|
|
8UZE
 
 | | Crystal structure of chimeric bat coronavirus BANAL-20-236 RBD complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of chimeric RBD complexed with chimeric mouse ACE2
To Be Published
|
|
8UF4
 
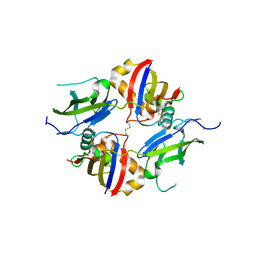 | | Crystal structure of wildtype dystroglycan proteolytic domain (juxtamembrane domain) | | Descriptor: | Beta-dystroglycan, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, M.J.M, Shi, K, Hayward, A.N, Uhlens, C, Evans III, R.L, Grant, E, Greenberg, L, Aihara, H, Gordon, W.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and Functional Analysis of Dystroglycan Cell Surface Cleavage Reveals a Novel Regulation Mechanism
To Be Published
|
|
8VQR
 
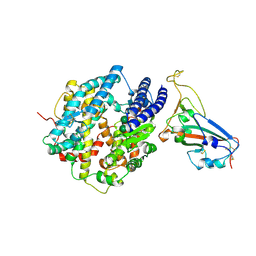 | | Crystal structure of chimeric SARS-CoV-2 RBD complexed with chimeric raccoon dog ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsueh, F.-C, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2024-01-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Structural basis for raccoon dog receptor recognition by SARS-CoV-2
To Be Published
|
|
8V9W
 
 | | X-ray crystal structure of JGFN4 complexed with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
9DK4
 
 | | ancestral hydroxynitrile lyase with fifteen substitutions | | Descriptor: | Fifteen-substitution variant of an ancestral hydroxynitrile lyase, SODIUM ION | | Authors: | Sarak, S.C, Tan, P, Evans III, R.L, Shi, K, Kazlauskas, R.J, Aihara, H, Pierce, C.T, Walsh, M.E, Greenberg, L.R. | | Deposit date: | 2024-09-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | To be published
To Be Published
|
|
4EP5
 
 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, SULFATE ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
7UFK
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UFL
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8V9X
 
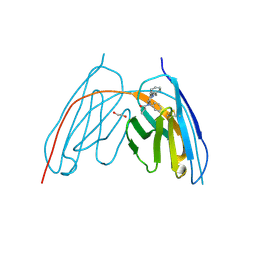 | | X-ray crystal structure of JGFN4 complex with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
4EP4
 
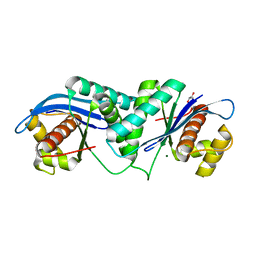 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, MAGNESIUM ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
8SNI
 
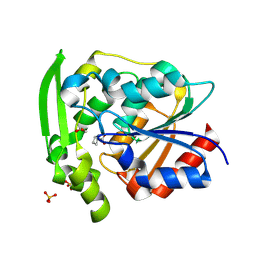 | | Hydroxynitrile Lyase from Hevea brasiliensis with Forty Mutations | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Walsh, M.E, Greenberg, L.R, Kazlauskas, R.J, Pierce, C.T, Aihara, H, Evans, R.L, Shi, K. | | Deposit date: | 2023-04-27 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be published
To Be Published
|
|
1P79
 
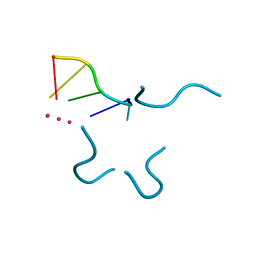 | | Crystal structure of a bulged RNA tetraplex: implications for a novel binding site in RNA tetraplex | | Descriptor: | 5'-R(*UP*GP*UP*GP*GP*U)-3', POTASSIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of a Bulged RNA Tetraplex at 1.1 A Resolution: Implications for a Novel Binding Site in RNA Tetraplex
STRUCTURE, 11, 2003
|
|
2GRB
 
 | |
2IP2
 
 | |
1MDG
 
 | | An Alternating Antiparallel Octaplex in an RNA Crystal Structure | | Descriptor: | 5'-R(*UP*(BGM)GP*AP*GP*GP*U)-3', COBALT HEXAMMINE(III), SODIUM ION | | Authors: | Pan, B.C, Xiong, Y, Shi, K, Sundaralingam, M. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Eight-Stranded Helical Fragment in RNA Crystal Structure: Implications for Tetraplex Interaction
Structure, 11, 2003
|
|
1FUF
 
 | | CRYSTAL STRUCTURE OF A 14BP RNA OLIGONUCLEOTIDE CONTAINING DOUBLE UU BULGES: A NOVEL INTRAMOLECULAR U*(AU) BASE TRIPLE | | Descriptor: | (5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3'), MAGNESIUM ION, SPERMINE | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2000-09-15 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
1J6S
 
 | | Crystal Structure of an RNA Tetraplex (UGAGGU)4 with A-tetrads, G-tetrads, U-tetrads and G-U octads | | Descriptor: | 5'-R(*(BRUP*GP*AP*GP*GP*U)-3', BARIUM ION, SODIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Deng, J, Sundaralingam, M. | | Deposit date: | 2002-07-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads:
implications for specific binding in RNA tetraplexes
Structure, 11, 2003
|
|
1IK5
 
 | | Crystal Structure of a 14mer RNA Containing Double UU Bulges in Two Crystal Forms: A Novel U*(AU) Intramolecular Base Triple | | Descriptor: | 5'-R(*GP*GP*UP*AP*UP*UP*UP*CP*GP*GP*UP*AP*(CBR)P*C)-3', 5'-R(*GP*GP*UP*AP*UP*UP*UP*UP*GP*GP*UP*AP*(CBR)P*C)-3', MAGNESIUM ION | | Authors: | Deng, J, Xiong, Y, Sudarsanakumar, C, Shi, K, Sundaralingam, M. | | Deposit date: | 2001-05-02 | | Release date: | 2001-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two forms of a 14-mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A x U) base triple.
RNA, 7, 2001
|
|
7JT4
 
 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | Descriptor: | Nucleosome-remodeling factor subunit BPTF | | Authors: | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-08-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5WS3
 
 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
7JN3
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
