6IU8
 
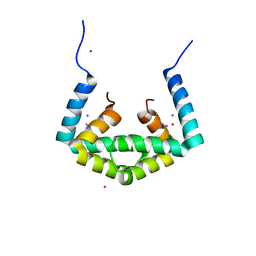 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
1MJJ
 
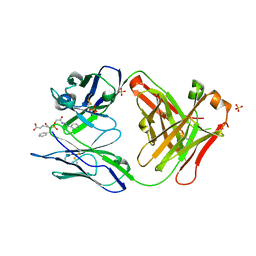 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE COMPLEX OF THE FAB FRAGMENT OF ESTEROLYTIC ANTIBODY MS6-12 AND A TRANSITION-STATE ANALOG | | Descriptor: | IMMUNOGLOBULIN MS6-12, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MH5
 
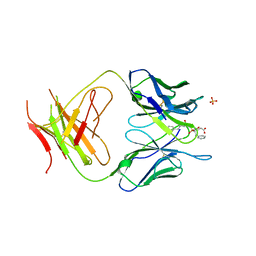 | | The Structure Of The Complex Of The Fab Fragment Of The Esterolytic Antibody MS6-164 and A Transition-State Analog | | Descriptor: | IMMUNOGLOBULIN MS6-164, N-{[2-({[1-(4-CARBOXYBUTANOYL)AMINO]-2-PHENYLETHYL}-HYDROXYPHOSPHINYL)OXY]ACETYL}-2-PHENYLETHYLAMINE, SULFATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MJ8
 
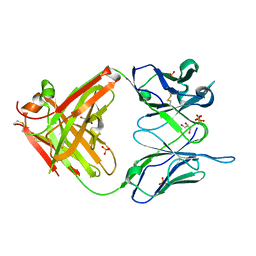 | | High Resolution Crystal Structure Of The Fab Fragment of The Esterolytic Antibody MS6-126 | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN MS6-126, PHOSPHATE ION | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-27 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
3MBE
 
 | | TCR 21.30 in complex with MHC class II I-Ag7HEL(11-27) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS II H2-IAg7 ALPHA CHAIN, ... | | Authors: | Corper, A.L, Yoshida, K, Teyton, L, Wilson, I.A. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | The diabetogenic mouse MHC class II molecule I-Ag7 is endowed with a switch that modulates TCR affinity.
J.Clin.Invest., 120, 2010
|
|
6IGL
 
 | | Crystal Structure of human ETB receptor in complex with IRL1620 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | Authors: | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
7S0N
 
 | | Structure of MS3494 from Mycobacterium Smegmatis determined by Solution NMR | | Descriptor: | Secreted protein | | Authors: | Kent, J.E, Tian, Y, Shin, K, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-08-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of MS3494 from Mycobacterium Smegmatis
To Be Published
|
|
8P3V
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 3 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3X
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Z
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Y
 
 | | Homomeric GluA2 flip R/G-edited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Krieger, J.M, Zhang, D, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
1F54
 
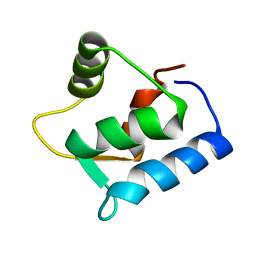 | | SOLUTION STRUCTURE OF THE APO N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1F55
 
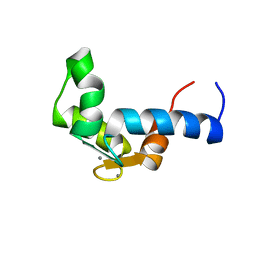 | | SOLUTION STRUCTURE OF THE CALCIUM BOUND N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
2RR2
 
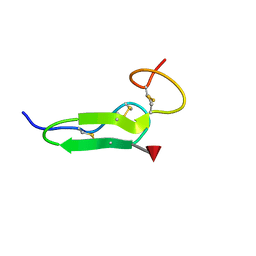 | | Structure of O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1, alpha-L-fucopyranose | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
6JT5
 
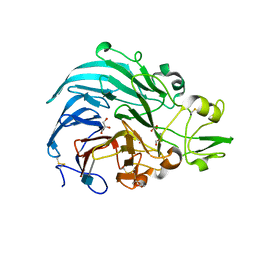 | | Crystal structure of PQQ doamin of Pyranose Dehydrogenase from Coprinopsis cinerea: apo-from | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular PQQ-dependent sugar dehydrogenase, ... | | Authors: | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | Deposit date: | 2019-04-09 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
7EK2
 
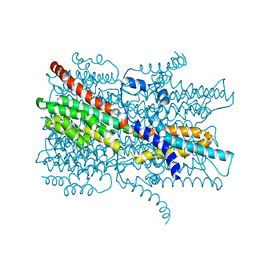 | | Cryo-EM structure of VCCN1 in lipid nanodisc | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
7EK1
 
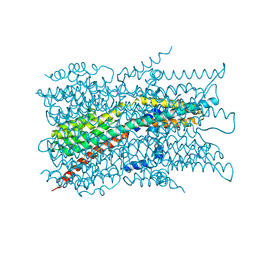 | | Cryo-EM structure of VCCN1 in detergent | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
7EK3
 
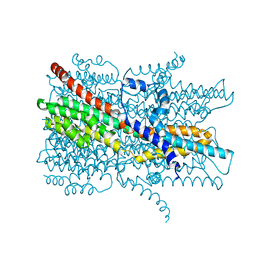 | | Cryo-EM structure of VCCN1 Y332A mutant in lipid nanodisc | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
2RR0
 
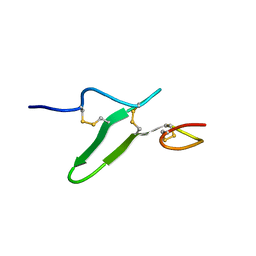 | | Structure of epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
6JWF
 
 | | Holo form of Pyranose Dehydrogenase PQQ domain from Coprinopsis cinerea | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | Deposit date: | 2019-04-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
1MIE
 
 | | Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 | | Descriptor: | IMMUNOGLOBULIN MS5-393 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-23 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
8HU1
 
 | | E. coli 70S ribosome complexed with tRNA_Ile2 bearing L34 and ct6A37 in classical state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-12-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nat.Struct.Mol.Biol., 31, 2024
|
|
