6BUM
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica | | Descriptor: | 1,3-PROPANDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shi, K, Cho, S, Seffernick, J.L, Bera, A, Wackett, L.P, Aihara, H. | | Deposit date: | 2017-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6BUO
 
 | |
6CWJ
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with 1,3-Acetone Dicarboxylic Acid | | Descriptor: | 1,3-PROPANDIOL, 3-oxopentanedioic acid, ACETATE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-03-30 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6BUP
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with cyanuric acid | | Descriptor: | 1,3,5-triazine-2,4,6-triol, 1,3-PROPANDIOL, CALCIUM ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2017-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6BUQ
 
 | |
6DHJ
 
 | |
6CA4
 
 | | Crystal structure of humanized D. rerio TDP2 by 14 mutations | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Shi, K, Aihrara, H. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | New fluorescence-based high-throughput screening assay for small molecule inhibitors of tyrosyl-DNA phosphodiesterase 2 (TDP2).
Eur J Pharm Sci, 118, 2018
|
|
6DRT
 
 | |
7K32
 
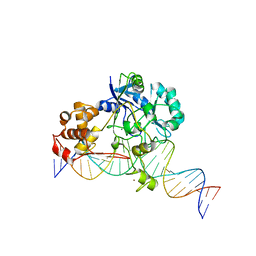 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K33
 
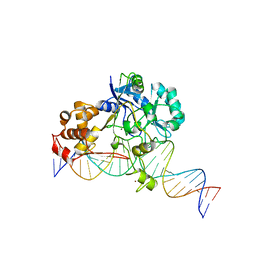 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3SG9
 
 | |
3SGC
 
 | |
3SG8
 
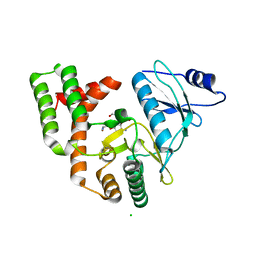 | |
2GRM
 
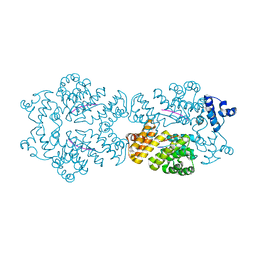 | | Crystal structure of PrgX/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
2GRL
 
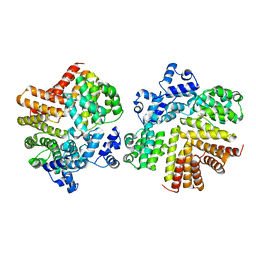 | | Crystal structure of dCT/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
1DVL
 
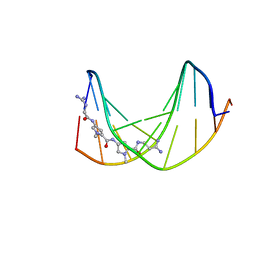 | |
438D
 
 | |
1JO2
 
 | |
1L3Z
 
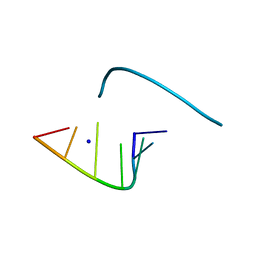 | | Crystal Structure Analysis of an RNA Heptamer | | Descriptor: | 5'-R(*GP*UP*AP*UP*AP*CP*A)-3', SODIUM ION | | Authors: | Shi, K, Pan, B, Sundaralingam, M. | | Deposit date: | 2002-03-04 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The crystal structure of an alternating RNA heptamer r(GUAUACA)
forming a six base-paired duplex with 3'-end adenine overhangs
Nucleic Acids Res., 31, 2003
|
|
6XR0
 
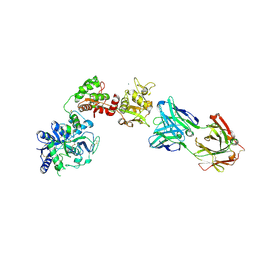 | | Crystal Structure of Human Melanotransferrin in complex with SC57.32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | Authors: | Hayashi, K, Longenecker, K.L, Vivona, S. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.064 Å) | | Cite: | Complex of human Melanotransferrin and SC57.32 Fab fragment reveals novel interdomain arrangement with ferric N-lobe and open C-lobe.
Sci Rep, 11, 2021
|
|
1MR8
 
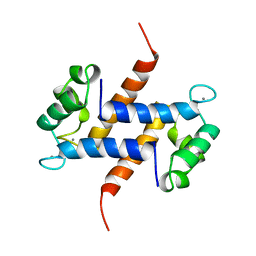 | | MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 FROM HUMAN | | Descriptor: | CALCIUM ION, MIGRATION INHIBITORY FACTOR-RELATED PROTEIN 8 | | Authors: | Ishikawa, K, Nakagawa, A, Tanaka, I, Nishihira, J. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of human MRP8, a member of the S100 calcium-binding protein family, by MAD phasing at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EOI
 
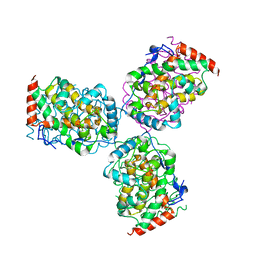 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE COMPLEXED WITH THE TRANSITION STATE ANALOG MOLYBDATE | | Descriptor: | ACID PHOSPHATASE, MOLYBDATE ION | | Authors: | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2001-03-23 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
7D56
 
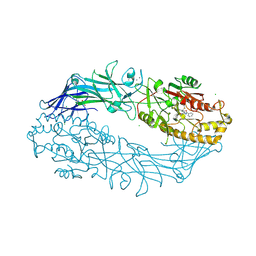 | |
7D8N
 
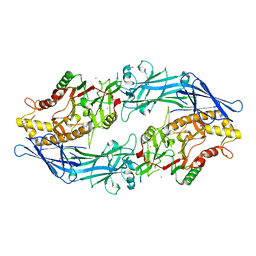 | | Structure of the inactive form of wild-type peptidylarginine deiminase type III (PAD3) crystallized under the condition with high concentrations of Ca2+ | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Funabashi, K, Sawata, M, Unno, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
6LPB
 
 | | Cryo-EM structure of the human PAC1 receptor coupled to an engineered heterotrimeric G protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Shihoya, W, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-11 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human PAC1 receptor coupled to an engineered heterotrimeric G protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
