6OGP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-063 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGT
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P51) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
3VL9
 
 | | Crystal structure of xeg-xyloglucan | | Descriptor: | Xyloglucan-specific endo-beta-1,4-glucanase A, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
6OGQ
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OYR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-002 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OGS
 
 | | X-ray crystal structure of darunavir-resistant HIV-1 protease (P30) in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
6OYD
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-004 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-14 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
3VLA
 
 | | Crystal structure of edgp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VLB
 
 | | Crystal structure of xeg-edgp | | Descriptor: | EDGP, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
6MCS
 
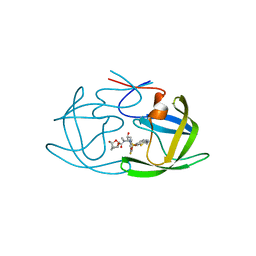 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6MCR
 
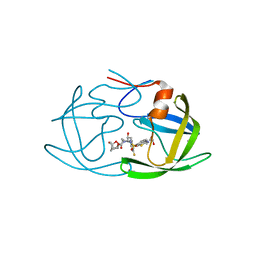 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
4TKB
 
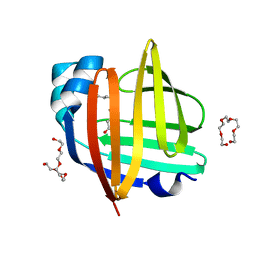 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lauric acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TJZ
 
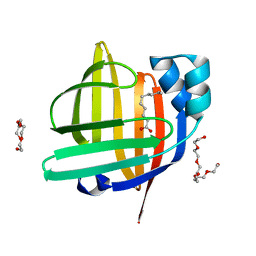 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with capric acid | | Descriptor: | DECANOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKH
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with myristic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKJ
 
 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XZG
 
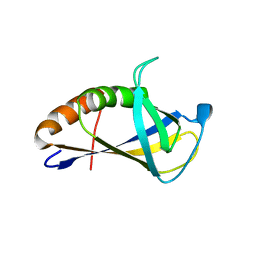 | | Crystal structure of HIRAN domain of human HLTF | | Descriptor: | Helicase-like transcription factor | | Authors: | Ikegaya, Y, Hara, K, Hashimoto, H. | | Deposit date: | 2015-02-04 | | Release date: | 2015-04-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance
J.Biol.Chem., 290, 2015
|
|
4DVZ
 
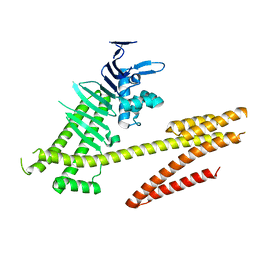 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
8JD8
 
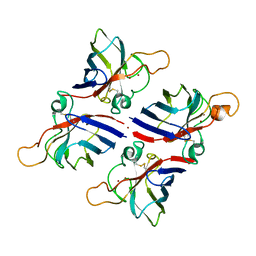 | | Crystal structure of Citrus limon Cu-Zn superoxide dismutase | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Utami, R.A, Yoshida, H, Retnoningrum, D.S, Ismaya, W.T. | | Deposit date: | 2023-05-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Direct relationship between dimeric form and activity in the acidic copper-zinc superoxide dismutase from lemon.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7Y8P
 
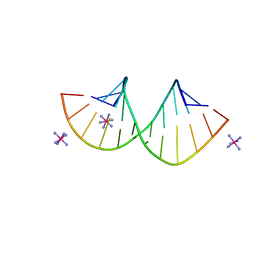 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
6MK9
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121
To Be Published
|
|
6MKL
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function analysis of darunavir-resistant-P51 HIV-1 protease in complex with GRL-142.
To Be Published
|
|
4ZJF
 
 | | Crystal structure of GP1 - the receptor binding domain of Lassa virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein | | Authors: | Cohen-Dvashi, H, Cohen, N, Israeli, H, Diskin, R. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Molecular Mechanism for LAMP1 Recognition by Lassa Virus.
J.Virol., 89, 2015
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6J
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
