8HN3
 
 | | Soluble domain of cytochrome c-556 from Chlorobaculum tepidum | | Descriptor: | ACETATE ION, Cytochrome c-556, GLYCEROL, ... | | Authors: | Kishimoto, H, Azai, C, Yamamoto, T, Mutoh, R, Nakaniwa, T, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
8HN2
 
 | | Selenomethionine-labelled soluble domain of Rieske iron-sulfur protein from chlorobaculum tepidum | | Descriptor: | Cytochrome b6-f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Kishimoto, H, Mutoh, R, Tanaka, H, Kurisu, G, Oh-oka, H. | | Deposit date: | 2022-12-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Soluble domains of cytochrome c-556 and Rieske iron-sulfur protein from Chlorobaculum tepidum: Crystal structures and interaction analysis.
Curr Res Struct Biol, 5, 2023
|
|
5T1F
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
5T1E
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
1IQC
 
 | | Crystal structure of Di-Heme Peroxidase from Nitrosomonas europaea | | Descriptor: | CALCIUM ION, GLYCEROL, HEME C, ... | | Authors: | Shimizu, H. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Nitrosomonas europaea cytochrome c peroxidase and the structural basis for ligand switching in bacterial di-heme peroxidases
Biochemistry, 40, 2001
|
|
3IEM
 
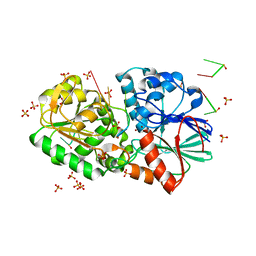 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog | | Descriptor: | CITRATE ANION, RNA (5'-R(*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU))-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitus, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with RNA analog
To be Published
|
|
6IZZ
 
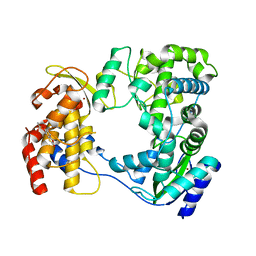 | |
3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEL
 
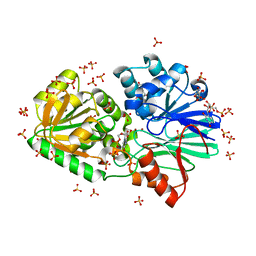 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
6J00
 
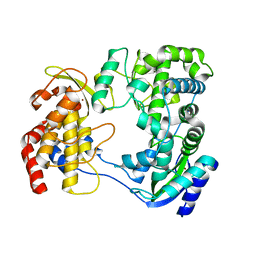 | |
3IE1
 
 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | Descriptor: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3IE2
 
 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IDZ
 
 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IE0
 
 | | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
6IZY
 
 | |
6IZX
 
 | | The RNA-dependent RNA polymerase domain of dengue 2 NS5, bound with RK-0404678 | | Descriptor: | 2-oxo-2H-1,3-benzoxathiol-5-yl acetate, COBALT (II) ION, Genome polyprotein, ... | | Authors: | Shimizu, H, Sekine, S. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase.
Plos Negl Trop Dis, 13, 2019
|
|
8HDD
 
 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Sode, K. | | Deposit date: | 2022-11-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
1NDH
 
 | |
2FAU
 
 | | Crystal structure of human vps26 | | Descriptor: | GLYCEROL, Vacuolar protein sorting 26 | | Authors: | Shi, H, Rojas, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2005-12-08 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The retromer subunit Vps26 has an arrestin fold and binds Vps35 through its C-terminal domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2RSD
 
 | | Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice | | Descriptor: | E3 SUMO-protein ligase SIZ1, ZINC ION | | Authors: | Shindo, H, Tsuchiya, W, Suzuki, R, Yamazaki, T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2
Febs Lett., 586, 2012
|
|
1KXW
 
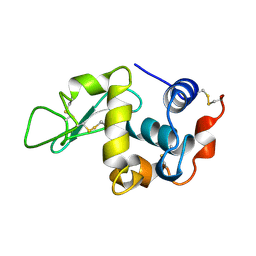 | |
1KXX
 
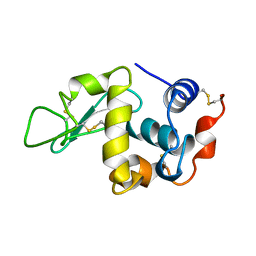 | |
1KXY
 
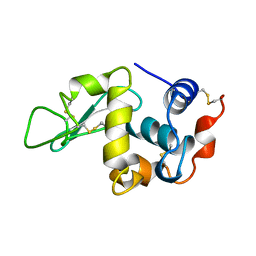 | |
6M73
 
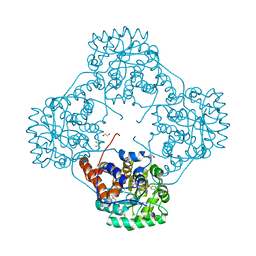 | | Crystal structure of Enterococcus hirae L-lactate oxidase in complex with D-lactate form ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Yoshida, H, Hiraka, K, Tsugawa, W, Sode, K. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of lactate oxidase from Enterococcus hirae revealed new aspects of active site loop function: Product-inhibition mechanism and oxygen gatekeeper
Protein Sci., 31, 2022
|
|
