3LMG
 
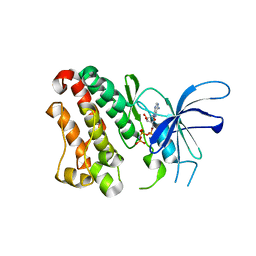 | | Crystal structure of the ERBB3 kinase domain in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Shi, F, Lemmon, M.A. | | Deposit date: | 2010-01-30 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8J72
 
 | | Crystal structure of mammalian Trim71 in complex with lncRNA Trincr1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM71, lncRNA Trincr1 | | Authors: | Shi, F.D, Zhang, K, Che, S.Y, Zhi, S.X, Yang, N. | | Deposit date: | 2023-04-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Molecular mechanism governing RNA-binding property of mammalian TRIM71 protein.
Sci Bull (Beijing), 69, 2024
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
8ROY
 
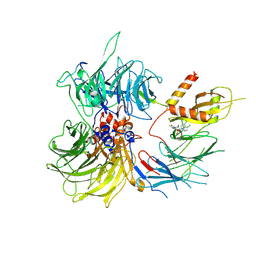 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROX
 
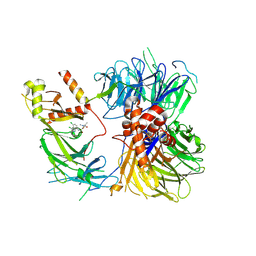 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
2RPZ
 
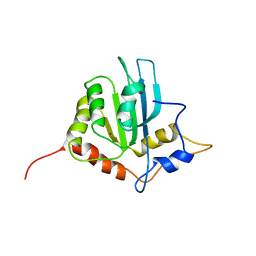 | | Solution structure of the monomeric form of mouse APOBEC2 | | Descriptor: | Probable C->U-editing enzyme APOBEC-2, ZINC ION | | Authors: | Hayashi, F, Nagata, T, Nagashima, T, Muto, Y, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-12-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monomeric form of mouse APOBEC2
To be Published
|
|
2CSW
 
 | |
2HR1
 
 | |
1UH6
 
 | | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06 | | Descriptor: | ubiquitin-like 5 | | Authors: | Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-25 | | Release date: | 2003-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the murine ubiquitin-like 5 protein from RIKEN cDNA 0610031K06
To be Published
|
|
3VJL
 
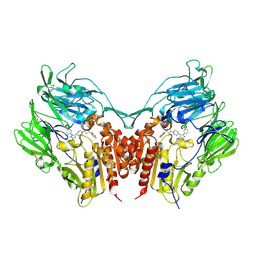 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3VJK
 
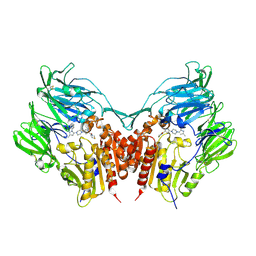 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with MP-513 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and preclinical profile of teneligliptin (3-[(2S,4S)-4-[4-(3-methyl-1-phenyl-1H-pyrazol-5-yl)piperazin-1-yl]pyrrolidin-2-ylcarbonyl]thiazolidine): A highly potent, selective, long-lasting and orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem., 20, 2012
|
|
3VJM
 
 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | Descriptor: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
2YUB
 
 | |
2YUC
 
 | |
1WLO
 
 | |
1WLN
 
 | |
2Z6A
 
 | |
2Z6U
 
 | |
2Z6Q
 
 | |
2ZCJ
 
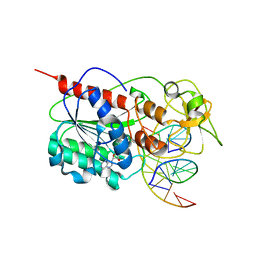 | | Ternary structure of the Glu119Gln M.HhaI, C5-Cytosine DNA methyltransferase, with unmodified DNA and AdoHcy | | Descriptor: | DNA (5'-D(*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), DNA (5'-D(*DTP*DGP*DAP*DTP*DAP*DGP*DCP*DGP*DCP*DTP*DAP*DTP*DC)-3'), Modification methylase HhaI, ... | | Authors: | Shieh, F.K, Reich, N.O. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | AdoMet-dependent Methyl-transfer: Glu(119) Is Essential for DNA C5-Cytosine Methyltransferase M.HhaI
J.Mol.Biol., 373, 2007
|
|
2B0T
 
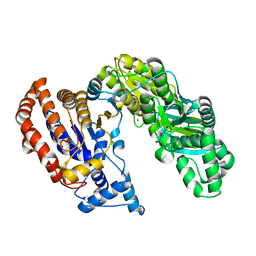 | | Structure of Monomeric NADP Isocitrate dehydrogenase | | Descriptor: | MAGNESIUM ION, NADP Isocitrate dehydrogenase | | Authors: | Imabayashi, F, Aich, S, Prasad, L, Delbaere, L.T. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-free structure of a monomeric NADP isocitrate dehydrogenase: An open conformation phylogenetic relationship of isocitrate dehydrogenase.
Proteins, 63, 2006
|
|
5XR9
 
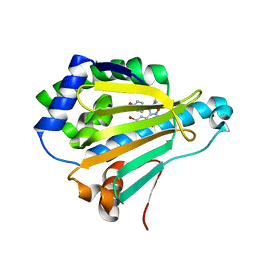 | | Crystal structure of Human Hsp90 with FS6 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-bromanyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Human Hsp90 with FS6
To Be Published
|
|
5XRE
 
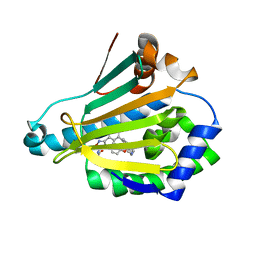 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[4-(4-methoxyphenyl)-3-[5-(8-methylquinolin-5-yl)-2,4-bis(oxidanyl)phenyl]-1,2-oxazol-5-yl]ethanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-08 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor JX1
To Be Published
|
|
5XQE
 
 | | Crystal structure of Human Hsp90 with FS3 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[3-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]-2-methyl-propanamide | | Authors: | Li, J, Shi, F, Xiong, B, He, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of Human Hsp90 with FS3
To Be Published
|
|
5XRD
 
 | |
