1GU9
 
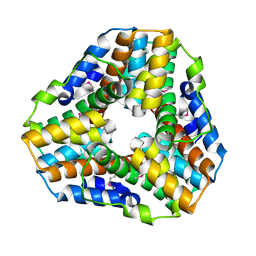 | | Crystal Structure of Mycobacterium tuberculosis Alkylperoxidase AhpD | | Descriptor: | ALKYLHYDROPEROXIDASE D | | Authors: | Nunn, C.M, Djordjevic, S, Hillas, P.J, Nishida, C, Ortiz de Montellano, P.R. | | Deposit date: | 2002-01-24 | | Release date: | 2002-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Alkylhydroperoxidase Ahpd, a Potential Target for Antitubercular Drug Design
J.Biol.Chem., 277, 2002
|
|
6L27
 
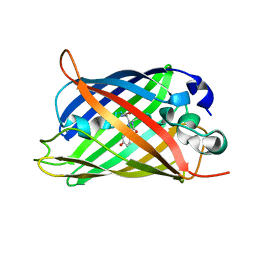 | | X-ray crystal structure of the mutant green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
6L26
 
 | | Neutron crystal structure of the mutant green fluorescent protein (EGFP) | | Descriptor: | Green fluorescent protein, trideuteriooxidanium | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-30 | | Method: | NEUTRON DIFFRACTION (1.444 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
1R12
 
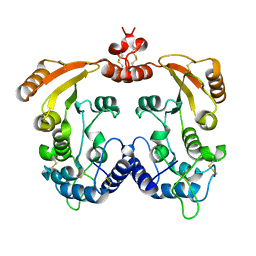 | | Native Aplysia ADP ribosyl cyclase | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1R0S
 
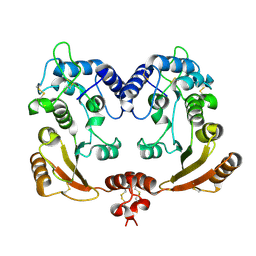 | | Crystal structure of ADP-ribosyl cyclase Glu179Ala mutant | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-22 | | Release date: | 2004-03-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
7BT2
 
 | | Crystal structure of the SERCA2a in the E2.ATP state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kabashima, Y, Ogawa, H, Nakajima, R, Toyoshima, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.00002861 Å) | | Cite: | What ATP binding does to the Ca2+pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7X7K
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Arg binding form | | Descriptor: | ARGININE, FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7J
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) L-Lys binding form | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
7X7I
 
 | | Ancestral L-Lys oxidase (AncLLysO-2) ligand free form | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Motoyama, T, Ishida, C, Hasebe, F, Ito, S, Nakano, S. | | Deposit date: | 2022-03-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Reaction Mechanism of Ancestral l-Lys alpha-Oxidase from Caulobacter Species Studied by Biochemical, Structural, and Computational Analysis
Acs Omega, 7, 2022
|
|
6LO9
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 6-cyclohexyloxy-9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86004949 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6LOB
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.40006661 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6LOA
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]-6-propan-2-yloxy-carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.50003314 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
7Y45
 
 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
7Y46
 
 | | Cryo-EM structure of the Na+,K+-ATPase in the E2.2K+ state after addition of ATP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Vilsen, B, Toyoshima, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Cryo-electron microscopy of Na + ,K + -ATPase reveals how the extracellular gate locks in the E2·2K + state.
Febs Lett., 596, 2022
|
|
6LOC
 
 | | Crystal structure of RORgammat with ligand C46D bound | | Descriptor: | 6-cyclobutyloxy-9-ethyl-~{N}-[(4-ethylsulfonylphenyl)methyl]carbazole-3-carboxamide, Nuclear receptor ROR-gamma | | Authors: | Feng, Y, Shijie, C. | | Deposit date: | 2020-01-04 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20014858 Å) | | Cite: | Crystal structure of RORgammat with ligand C46D bound
To Be Published
|
|
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
6T9O
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(3,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
4YCL
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ogawa, H, Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6A28
 
 | | Crystal structure of PprA W183R mutant form 2 | | Descriptor: | DNA repair protein PprA, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
6A27
 
 | | Crystal structure of PprA W183R mutant form 1 | | Descriptor: | DNA repair protein PprA, GLYCEROL, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.353 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
6A29
 
 | | Crystal structure of PprA A139R mutant | | Descriptor: | DNA repair protein PprA | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
