3AH1
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylneuramic acid | | Descriptor: | Main hemagglutinin component, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
2YUT
 
 | |
2YV4
 
 | |
2Z0G
 
 | | The crystal structure of PII protein | | Descriptor: | CHLORIDE ION, Nitrogen regulatory protein P-II, PHOSPHATE ION | | Authors: | Sakai, H, Shinkai, A, Kitamura, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of PII protein
To be Published
|
|
2ZDT
 
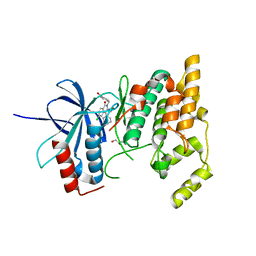 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(6-chloro-1-oxo-4-phenyl-3-propanoylisoquinolin-2(1H)-yl)methyl]benzoic acid, GLYCEROL, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Asano, Y, Fukumoto, S, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (2)
Bioorg.Med.Chem., 16, 2008
|
|
3ANP
 
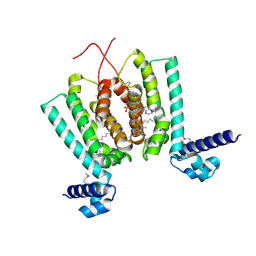 | | Crystal structure of Thermus thermophilus FadR, a TetR familly transcriptional repressor, in complex with lauroyl-CoA. | | Descriptor: | DODECYL-COA, LAURIC ACID, Transcriptional repressor, ... | | Authors: | Agari, Y, Agari, K, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
2Z1K
 
 | | Crystal Structure of Ttha1563 from Thermus thermophilus HB8 | | Descriptor: | (Neo)pullulanase, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), PHOSPHATE ION, ... | | Authors: | Niwa, H, Shimada, A, Matsunaga, E, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Ttha1563 from Thermus thermophilus HB8
To be Published
|
|
3ANG
 
 | | Crystal structure of Thermus thermophilus FadR in complex with E. coli-derived dodecyl-CoA | | Descriptor: | DODECYL-COA, Transcriptional repressor, TetR family | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-01 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
3BOY
 
 | | Crystal structure of the HutP antitermination complex bound to the HUT mRNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*UP*UP*UP*AP*GP*UP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Balasundaresan, D, Jeyakanthan, J, Shinkai, A, Yokoyama, S, Kumar, P.K.R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of HutP complexed with the 55-mer RNA
To be Published
|
|
2ZCA
 
 | | Crystal Structure of TTHB189, a CRISPR-associated protein, Cse2 family from Thermus thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB189 | | Authors: | Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-08 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
3AAI
 
 | |
2ZZQ
 
 | | Crystal structure analysis of the HEV capsid protein, PORF2 | | Descriptor: | Protein ORF3, Capsid protein | | Authors: | Miyazaki, N, Xing, L, Wang, C.-Y, Li, T.-C, Takeda, N, Higashiura, A, Nakagawa, A, Tsukihara, T, Miyamura, T, Cheng, R.H. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Role of protein domain-modularity in designating capsid assembly and antigenicity
To be Published
|
|
2ZDU
 
 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(4-{[6-bromo-3-(methoxycarbonyl)-1-oxo-4-phenylisoquinolin-2(1H)-yl]methyl}phenyl)amino]-4-oxobutanoic acid, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Ohra, T, Itoh, F, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (1)
Bioorg.Med.Chem., 16, 2008
|
|
2EB7
 
 | | Crystal structure of the hypothetical regulator from Sulfolobus tokodaii 7 | | Descriptor: | 146aa long hypothetical transcriptional regulator | | Authors: | Kumarevel, T.S, Nishio, M, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the MarR family regulatory protein, ST1710, from Sulfolobus tokodaii strain 7
J.Struct.Biol., 161, 2008
|
|
2D60
 
 | | Crystal structure of deoxy human hemoglobin complexed with two L35 molecules | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, Hemoglobin alpha subunit, Hemoglobin beta subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2D5X
 
 | | Crystal structure of carbonmonoxy horse hemoglobin complexed with L35 | | Descriptor: | 2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID, CARBON MONOXIDE, Hemoglobin alpha subunit, ... | | Authors: | Yokoyama, T, Neya, S, Tsuneshige, A, Yonetani, T, Park, S.Y, Tame, J.R. | | Deposit date: | 2005-11-08 | | Release date: | 2006-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | R-state haemoglobin with low oxygen affinity: crystal structures of deoxy human and carbonmonoxy horse haemoglobin bound to the effector molecule L35
J.Mol.Biol., 356, 2006
|
|
2DJW
 
 | | Crystal structure of TTHA0845 from Thermus thermophilus HB8 | | Descriptor: | ZINC ION, probable transcriptional regulator, AsnC family | | Authors: | Okazaki, N, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-05 | | Release date: | 2006-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the stand-alone RAM-domain protein from Thermus thermophilus HB8
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2BFH
 
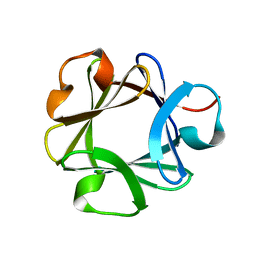 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
2CG5
 
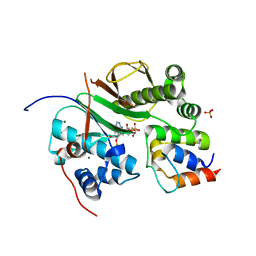 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase in complex with cytosolic acyl carrier protein and coenzyme A | | Descriptor: | COENZYME A, FATTY ACID SYNTHASE, L-AMINOADIPATE-SEMIALDEHYDE DEHYDROGENASE-PHOSPHOPANTETHEINYL TRANSFERASE, ... | | Authors: | Bunkoczi, G, Joshi, A, Papagrigoriu, E, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Smith, S, Oppermann, U. | | Deposit date: | 2006-02-27 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and Substrate Recognition of Human Holo Acp Synthase.
Chem.Biol., 14, 2007
|
|
2EG4
 
 | | Crystal Structure of Probable Thiosulfate Sulfurtransferase | | Descriptor: | Probable thiosulfate sulfurtransferase, SULFATE ION, ZINC ION | | Authors: | Sakai, H, Ebihara, A, Kitamura, Y, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Probable Thiosulfate Sulfurtransferase
To be Published
|
|
2EBS
 
 | | Crystal Structure Anaalysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) D465N Mutant Complexed with a Xyloglucan Heptasaccharide | | Descriptor: | Oligoxyloglucan reducing end-specific cellobiohydrolase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yaoi, K, Kondo, H, Hiyoshi, A, Noro, N, Sugimoto, H, Miyazaki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structural Basis for the Exo-mode of Action in GH74 Oligoxyloglucan Reducing End-specific Cellobiohydrolase.
J.Mol.Biol., 370, 2007
|
|
2E85
 
 | | Crystal Structure of the Hydrogenase 3 Maturation protease | | Descriptor: | CALCIUM ION, Hydrogenase 3 maturation protease | | Authors: | Tanaka, T, Kumarevel, T.S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-18 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of hydrogenase maturating endopeptidase HycI from Escherichia coli
Biochem.Biophys.Res.Commun., 389, 2009
|
|
2EG3
 
 | | Crystal Structure of Probable Thiosulfate Sulfurtransferase | | Descriptor: | Probable thiosulfate sulfurtransferase, SULFATE ION, ZINC ION | | Authors: | Sakai, H, Ebihara, A, Kitamura, Y, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-27 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Probable Thiosulfate Sulfurtransferase
To be Published
|
|
2EQA
 
 | | Crystal Structure of the hypothetical Sua5 protein from Sulfolobus tokodaii | | Descriptor: | ADENOSINE MONOPHOSPHATE, Hypothetical protein ST1526, MAGNESIUM ION | | Authors: | Agari, Y, Shinkai, A, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7
Proteins, 70, 2008
|
|
