6XAL
 
 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Val) | | Descriptor: | (3S,6S)-3-[(1H-indol-3-yl)methyl]-6-(propan-2-yl)piperazine-2,5-dione, NzeB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAK
 
 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Pro) and cyclo-(L-Trp-L-Trp) | | Descriptor: | (3S,6S)-3,6-bis[(1H-indol-3-yl)methyl]piperazine-2,5-dione, (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAM
 
 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-homoalanine) | | Descriptor: | (3S,6S)-3-ethyl-6-[(1H-indol-3-yl)methyl]piperazine-2,5-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
3LCR
 
 | | Thioesterase from Tautomycetin Biosynthhetic Pathway | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Tautomycetin biosynthetic PKS | | Authors: | Akey, D.L, Scaglione, J.B, Smith, J.L, Sherman, D.H. | | Deposit date: | 2010-01-11 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the tautomycetin thioesterase: analysis of a stereoselective polyketide hydrolase.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6PVF
 
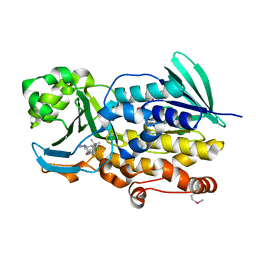 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVH
 
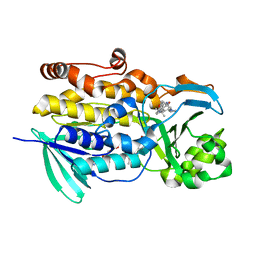 | | Crystal structure of PhqK in complex with paraherquamide K | | Descriptor: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVI
 
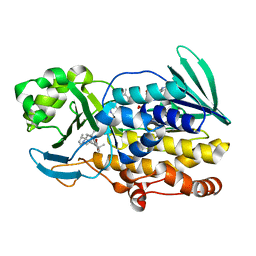 | | Crystal structure of PhqK in complex with paraherquamide L | | Descriptor: | (8aS,13S,13aR,14aS)-4,4,13,15,15-pentamethyl-12,13,14,14a,15,16-hexahydro-4H,8H,9H,11H-8a,13a-(epiminomethano)[1,4]dioxepino[2,3-a]indolizino[6,7-h]carbazol-17-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6PVJ
 
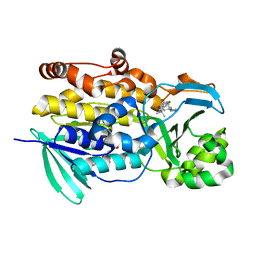 | | Crystal structure of PhqK in complex with malbrancheamide C | | Descriptor: | (5aS,12aS,13aS)-9-bromo-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
2VZ7
 
 | | Crystal structure of the YC-17-bound PikC D50N mutant | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, S, Sherman, D.H, Podust, L.M. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Analysis of Transient and Catalytic Desosamine Binding Pockets in Cytochrome P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 284, 2009
|
|
1KMZ
 
 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | Descriptor: | mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
1KLL
 
 | | Molecular basis of mitomycin C resictance in streptomyces: Crystal structures of the MRD protein with and without a drug derivative | | Descriptor: | 1,2-CIS-1-HYDROXY-2,7-DIAMINO-MITOSENE, mitomycin-binding protein | | Authors: | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | Deposit date: | 2001-12-12 | | Release date: | 2002-07-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
3F5H
 
 | | Crystal structure of fused docking domains from PikAIII and PikAIV of the pikromycin polyketide synthase | | Descriptor: | SODIUM ION, Type I polyketide synthase PikAIII, Type I polyketide synthase PikAIV fusion protein | | Authors: | Buchholz, T.J, Geders, T.W, Bartley, F.E, Reynolds, K.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains.
Acs Chem.Biol., 4, 2009
|
|
5WIS
 
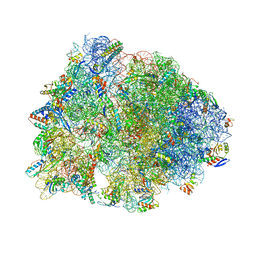 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with methymycin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
5WIT
 
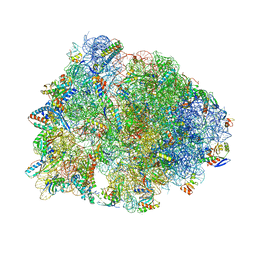 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pikromycin and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | (3R,5R,6S,7S,9R,11E,13S,14R)-14-ethyl-13-hydroxy-3,5,7,9,13-pentamethyl-2,4,10-trioxo-1-oxacyclotetradec-11-en-6-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Almutairi, M.M, Svetlov, M.S, Hansen, D.A, Khabibullina, N.F, Klepacki, D, Kang, H.Y, Sherman, D.H, Vazquez-Laslop, N, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2017-07-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins.
Nucleic Acids Res., 45, 2017
|
|
7S3J
 
 | | Crystal Structure of AspB P450 in complex with brevianamide F substrates | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, AspB, GLYCEROL, ... | | Authors: | Newmister, S.A, Shende, V.V, Harris, N.R, Sanders, J.N, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-07 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1ZLK
 
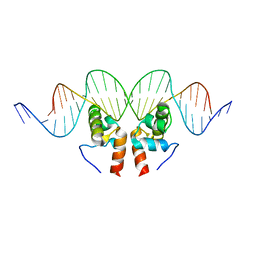 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR C-terminal Domain-DNA Complex | | Descriptor: | 5'-D(*CP*GP*TP*GP*GP*CP*CP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*AP*CP*TP*TP*TP*AP*GP*TP*CP*CP*CP*CP*AP*AP*AP*GP*CP*GP*CP*GP*GP*GP*CP*CP*AP*T)-3', 5'-D(*GP*GP*CP*CP*CP*GP*CP*GP*CP*TP*TP*TP*GP*GP*GP*GP*AP*CP*TP*AP*AP*AP*GP*TP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*GP*GP*CP*CP*AP*CP*GP*AP*T)-3', Dormancy Survival Regulator | | Authors: | Wisedchaisri, G, Wu, M, Rice, A.E, Roberts, D.M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency.
J.Mol.Biol., 354, 2005
|
|
3DX5
 
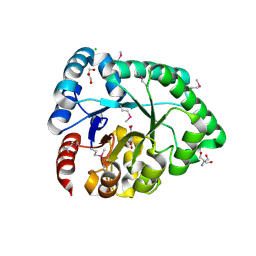 | | Crystal structure of the probable 3-DHS dehydratase AsbF involved in the petrobactin synthesis from Bacillus anthracis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stols, L, Eschenfeldt, W, Pfleger, B.F, Sherman, D.H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-23 | | Release date: | 2008-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and functional analysis of AsbF: origin of the stealth 3,4-dihydroxybenzoic acid subunit for petrobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3FWO
 
 | | The large ribosomal subunit from Deinococcus radiodurans complexed with Methymycin | | Descriptor: | (3R,4S,5S,7R,9E,11S,12R)-12-ethyl-11-hydroxy-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S RIBOSOMAL RNA, 5S RIBOSOMAL RNA | | Authors: | Auerbach, T, Mermershtain, I, Bashan, A, Davidovich, C, Rozenberg, H, Sherman, D.H, Yonath, A. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for the antibacterial activity of the 12-membered-ring mono-sugar macrolide methymycin
Biotechnologia, 1, 2009
|
|
7UF8
 
 | | Structure of CtdP in complex with penicimutamide E and NADP+ | | Descriptor: | 1,2-ETHANEDIOL, CtdP, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rivera, S, Liu, Z, Newmister, S.A, Gao, X, Sherman, D.H. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An NmrA-like enzyme-catalysed redox-mediated Diels-Alder cycloaddition with anti-selectivity.
Nat.Chem., 15, 2023
|
|
6XA2
 
 | | Structure of the tirandamycin C-bound P450 monooxygenase TamI | | Descriptor: | (3E)-3-{(2E,4E,6R)-1-hydroxy-4-methyl-6-[(1R,3R,4S,5R)-1,4,8-trimethyl-2,9-dioxabicyclo[3.3.1]non-7-en-3-yl]hepta-2,4-dien-1-ylidene}-2H-pyrrole-2,4(3H)-dione, PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
6XAI
 
 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-Pro) | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
6XAJ
 
 | | Crystal structure of NzeB | | Descriptor: | 1,2-ETHANEDIOL, NzeB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
3RIX
 
 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
2WHW
 
 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLOTRIDECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WI9
 
 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLODODECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
