6E9T
 
 | | DHF58 filament | | Descriptor: | DHF58 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9Y
 
 | | DHF38 filament | | Descriptor: | DHF38 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9X
 
 | | DHF91 filament | | Descriptor: | DHF91 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
5J8J
 
 | | A histone deacetylase from Saccharomyces cerevisiae | | Descriptor: | Histone deacetylase HDA1 | | Authors: | Zhu, Y, Shen, H, Li, X, Teng, M. | | Deposit date: | 2016-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Structural and histone binding ability characterization of the ARB2 domain of a histone deacetylase Hda1 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
8K14
 
 | | X-ray crystal structure of 18a in BRD4(1) | | Descriptor: | 4-[8-methoxy-2-methyl-1-(1-phenylethyl)imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Xu, H, Shen, H, Zhang, Y, Xu, Y, Li, R. | | Deposit date: | 2023-07-10 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of (R)-4-(8-methoxy-2-methyl-1-(1-phenylethy)-1H-imidazo[4,5-c]quinnolin-7-yl)-3,5-dimethylisoxazole as a potent and selective BET inhibitor for treatment of acute myeloid leukemia (AML) guided by FEP calculation.
Eur.J.Med.Chem., 263, 2024
|
|
7VLA
 
 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
6E9R
 
 | | DHF46 filament | | Descriptor: | DHF46 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
1IL5
 
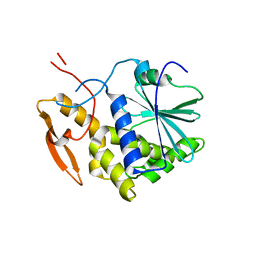 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 2,5-DIAMINO-4,6-DIHYDROXYPYRIMIDINE (DDP) | | Descriptor: | 2,4-DIAMINO-4,6-DIHYDROXYPYRIMIDINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL3
 
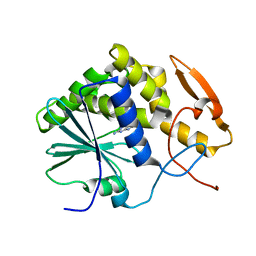 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 7-DEAZAGUANINE | | Descriptor: | 7-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL9
 
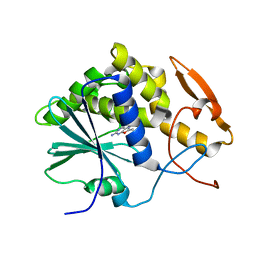 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 8-METHYL-9-OXOGUANINE | | Descriptor: | 5-AMINO-2-METHYL-6H-OXAZOLO[5,4-D]PYRIMIDIN-7-ONE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL4
 
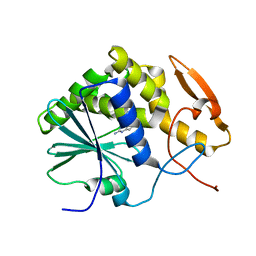 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 9-DEAZAGUANINE | | Descriptor: | 9-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
3FKS
 
 | | Yeast F1 ATPase in the absence of bound nucleotides | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Kabaleeswaran, V, Symersky, J, Shen, H, Walker, J.E, Leslie, A.G.W, Mueller, D.M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.587 Å) | | Cite: | Asymmetric structure of the yeast f1 ATPase in the absence of bound nucleotides.
J.Biol.Chem., 284, 2009
|
|
7YTD
 
 | | Cryo-EM structure of four human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YSG
 
 | | Cryo-EM structure of human FcmR bound to sIgM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
7YTE
 
 | |
7YTC
 
 | | Cryo-EM structure of human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
8WY7
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 4 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WY3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 21 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-cyclopropyl-2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]ethanamide | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WYG
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor 22 | | Descriptor: | 2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]-~{N}-(4-oxidanylcyclohexyl)ethanamide, Bromodomain-containing protein 2 | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8WXY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 23 | | Descriptor: | 5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-4-[(2-morpholin-4-yl-2-oxidanylidene-ethyl)amino]pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
8UBG
 
 | | DpHF19 filament | | Descriptor: | DpHF19,Green fluorescent protein (Fragment) | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
