8ESF
 
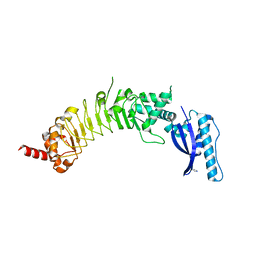 | |
3KEX
 
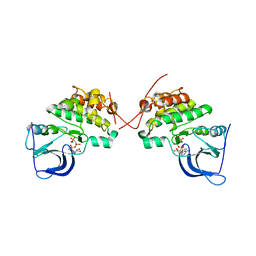 | | Crystal structure of the catalytically inactive kinase domain of the human epidermal growth factor receptor 3 (HER3) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Jura, N, Shan, Y, Cao, X, Shaw, D.E, Kuriyan, J. | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6MXT
 
 | | Crystal structure of human beta2 adrenergic receptor bound to salmeterol and Nb71 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endolysin, ... | | Authors: | Masureel, M, Zou, Y, Picard, L.P, van der Westhuizen, E, Mahoney, J.P, Rodrigues, J.P.G.L.M, Mildorf, T.J, Dror, R.O, Shaw, D.E, Bouvier, M, Pardon, E, Steyaert, J, Sunahara, R.K, Weis, W.I, Zhang, C, Kobilka, B.K. | | Deposit date: | 2018-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95934224 Å) | | Cite: | Structural insights into binding specificity, efficacy and bias of a beta2AR partial agonist.
Nat. Chem. Biol., 14, 2018
|
|
3LH9
 
 | |
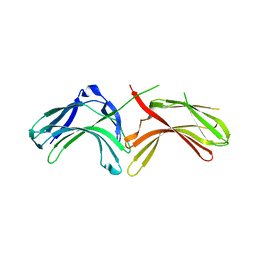
3LH8
 
 | |
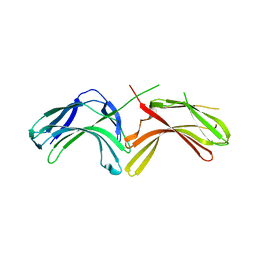
3LHA
 
 | |
3G6H
 
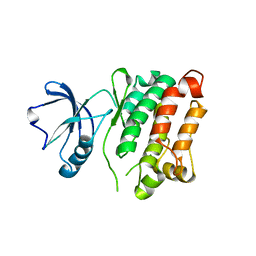 | | Src Thr338Ile inhibited in the DFG-Asp-Out conformation | | Descriptor: | N-{4-methyl-3-[(3-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
3PSN
 
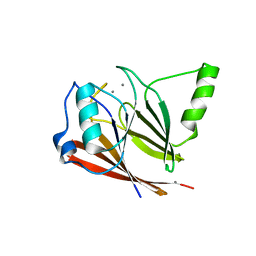 | | Crystal structure of mouse VPS29 complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3PSO
 
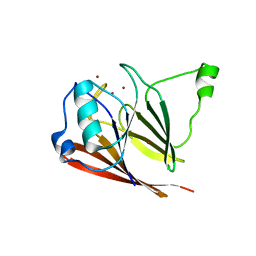 | | Crystal structure of mouse VPS29 complexed with Zn2+ | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
2M20
 
 | | EGFR transmembrane - juxtamembrane (TM-JM) segment in bicelles: MD guided NMR refined structure. | | Descriptor: | Epidermal growth factor receptor | | Authors: | Endres, N.F, Das, R, Smith, A, Arkhipov, A, Kovacs, E, Huang, Y, Pelton, J.G, Shan, Y, Shaw, D.E, Wemmer, D.E, Groves, J.T, Kuriyan, J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor.
Cell(Cambridge,Mass.), 152, 2013
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
1QQQ
 
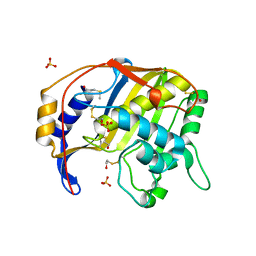 | | CRYSTAL STRUCTURE ANALYSIS OF SER254 MUTANT OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Fantz, C, Shaw, D, Jennings, W, Forsthoefel, A, Kitchens, M, Phan, J, Minor, W, Lebioda, L, Berger, F.G, Spencer, H.T. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Drug-resistant variants of Escherichia coli thymidylate synthase: effects of substitutions at Pro-254.
Mol.Pharmacol., 57, 2000
|
|
4DAJ
 
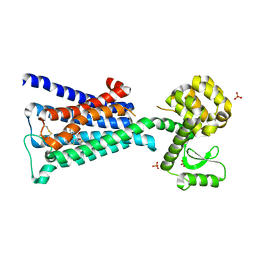 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
8G69
 
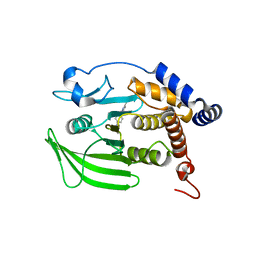 | | Wildtype PTP1b in complex with DES5743 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(5-ethyl-3-methyl-1H-pyrazol-1-yl)aniline, MAGNESIUM ION, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G67
 
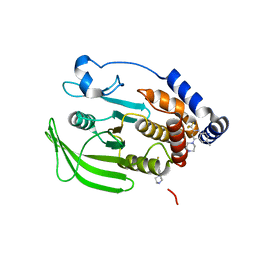 | | Wildtype PTP1b in complex with DES4884 | | Descriptor: | 6-methyl-4-(piperazin-1-yl)-2-(trifluoromethyl)quinoline, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G6A
 
 | | Wildtype PTP1b in complex with DES6016 | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G65
 
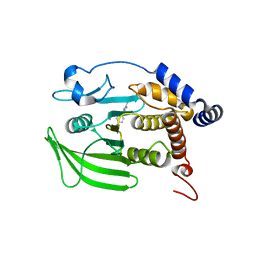 | | Wildtype PTP1b in complex with DES4799 | | Descriptor: | 4-(3,5-dimethyl-1H-pyrazol-1-yl)aniline, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
8G68
 
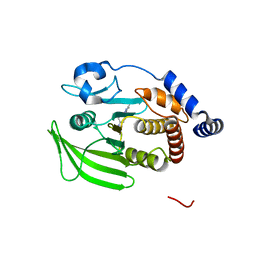 | | Wildtype PTP1b in complex with DES5742 | | Descriptor: | 4-(3-ethyl-5-methyl-1H-pyrazol-1-yl)aniline, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Willmore, L, Yeh, C.Y, Giordanetto, F, Shahamadtar, S, Nisonoff, H, Maragakis, P, Shaw, D.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and Validation of the Binding Poses of Allosteric Fragment Hits to Protein Tyrosine Phosphatase 1b: From Molecular Dynamics Simulations to X-ray Crystallography.
J.Chem.Inf.Model., 63, 2023
|
|
3G6G
 
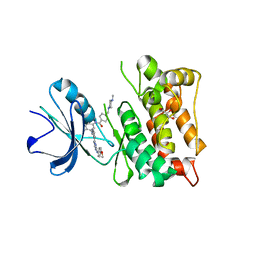 | | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations | | Descriptor: | GLYCEROL, N-{3-[(3-{4-[(4-methoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]-4-methylphenyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
1E41
 
 | |
1E3Y
 
 | |
2C7D
 
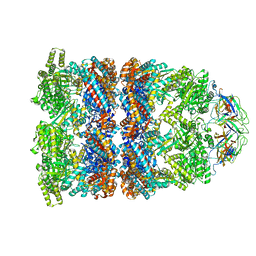 | | Fitted coordinates for GroEL-ADP7-GroES Cryo-EM complex (EMD-1181) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2C7C
 
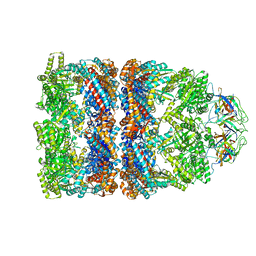 | | FITTED COORDINATES FOR GROEL-ATP7-GROES CRYO-EM COMPLEX (EMD-1180) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
