7SHX
 
 | |
7MDY
 
 | | LolCDE nucleotide-bound | | Descriptor: | ADP ORTHOVANADATE, Lipo-releasing system transmembrane protein lolC, Lipoprotein transporter subunit LolE, ... | | Authors: | Sharma, S, Liao, M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins
Nat Commun, 12, 2021
|
|
7MDX
 
 | | LolCDE nucleotide-free | | Descriptor: | (2R)-2-(tridecanoyloxy)propyl hexadecanoate, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, ... | | Authors: | Sharma, S, Liao, M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins
Nat Commun, 12, 2021
|
|
6U79
 
 | |
6W3M
 
 | |
8U1E
 
 | |
1NKX
 
 | | CRYSTAL STRUCTURE OF A PROTEOLYTICALLY GENERATED FUNCTIONAL MONOFERRIC C-LOBE OF BOVINE LACTOFERRIN AT 1.9A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Sharma, S, Jasti, J, Kumar, J, Mohanty, A.K, Singh, T.P. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Proteolytically Generated Functional Monoferric C-lobe of
Bovine Lactoferrin at 1.9A Resolution
J.Mol.Biol., 331, 2003
|
|
8F39
 
 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F29
 
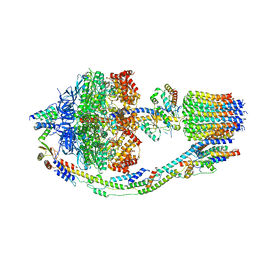 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FKJ
 
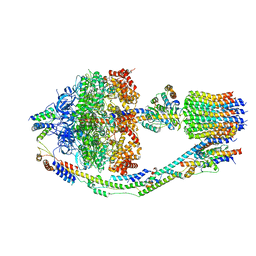 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FL8
 
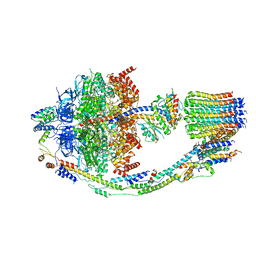 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2YIM
 
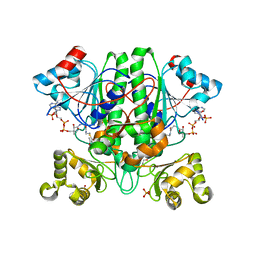 | | The enolisation chemistry of a thioester-dependent racemase: the 1.4 A crystal structure of a complex with a planar reaction intermediate analogue | | Descriptor: | 2-METHYLACETOACETYL COA, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sharma, S, Bhaumik, P, Venkatesan, R, Hiltunen, J.K, Conzelmann, E, Juffer, A.H, Wierenga, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The Enolization Chemistry of a Thioester-Dependent Racemase: The 1.4 A Crystal Structure of a Reaction Intermediate Complex Characterized by Detailed Qm/Mm Calculations.
J Phys Chem B, 116, 2012
|
|
6IF9
 
 | |
2MDL
 
 | |
2A7T
 
 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | Descriptor: | Neurotoxin | | Authors: | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-07-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
8ZN1
 
 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
8ZN4
 
 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
3C2X
 
 | | Crystal structure of peptidoglycan recognition protein at 1.8A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein, ... | | Authors: | Sharma, P, Singh, N, Sinha, M, Sharma, S, Perbandt, M, Betzel, C, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-01-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the peptidoglycan recognition protein at 1.8 A resolution reveals dual strategy to combat infection through two independent functional homodimers
J.Mol.Biol., 378, 2008
|
|
8ZOZ
 
 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
7XU8
 
 | | Structure of the complex of camel peptidoglycan recognition protein-short (PGRP-S) with heptanoic acid at 2.15 A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Maurya, A, Ahmad, N, Viswanathan, V, Singh, P.K, Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 angstrom resolution.
Int J Biochem Mol Biol, 13, 2022
|
|
7WYJ
 
 | | Structure of the complex of lactoperoxidase with nitric oxide catalytic product nitrite at 1.89 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Singh, R.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural evidence of the conversion of nitric oxide (NO) to nitrite ion (NO2-) by lactoperoxidase (LPO): Structure of the complex of LPO with NO2- at 1.89 angstrom resolution
J.Inorg.Biochem., 247, 2023
|
|
8ZN2
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.65 A resolution.
To Be Published
|
|
8ZN3
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.41 A resolution.
To Be Published
|
|
8JUK
 
 | |
6KY7
 
 | | Crystal structure of yak lactoperoxidase at 2.27 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 2.27 A resolution
To Be Published
|
|
