6MTD
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTE
 
 | | Rabbit 80S ribosome with eEF2 and SERBP1 (rotated state) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
3JAH
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JAG
 
 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JAI
 
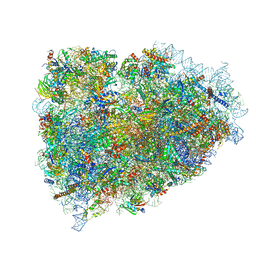 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
6WAM
 
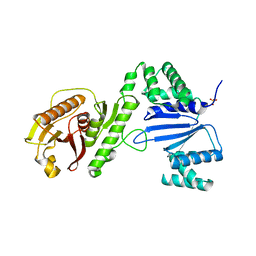 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6VM5
 
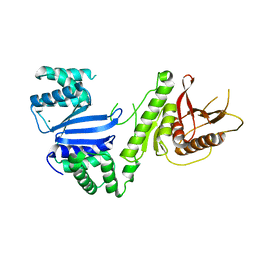 | | Structure of Moraxella osloensis Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6VM6
 
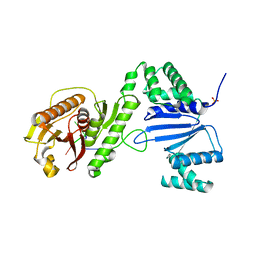 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 2'3'3'-cAAA | | Descriptor: | 2'-5'-Linked Cyclic RNA (5'-R(P*AP*AP*A)-3'), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6WAN
 
 | | Structure of Acinetobacter baumannii Cap4 SAVED/CARF-domain containing receptor with the cyclic trinucleotide 3'3'3'-cAAA | | Descriptor: | Cyclic RNA (R(P*AP*AP*A), SAVED domain-containing protein, SULFATE ION | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
7UN8
 
 | | SfSTING with c-di-GMP single fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UN9
 
 | | SfSTING with c-di-GMP double fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UNA
 
 | | SfSTING with cGAMP (masked) | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7VIM
 
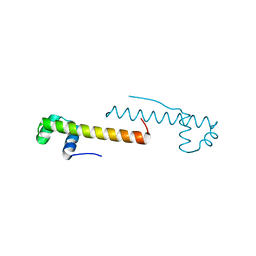 | |
7UN6
 
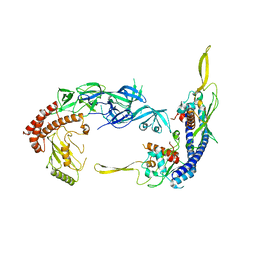 | | Complex of UBE2O with NAP1L1 | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme, Nucleosome assembly protein 1-like 1 | | Authors: | Yip, M.C.J, Sedor, S.F, Shao, S. | | Deposit date: | 2022-04-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of client selection by the protein quality-control factor UBE2O.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UN3
 
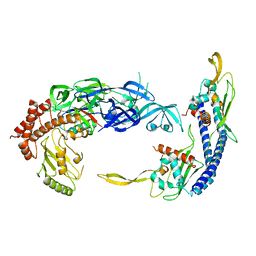 | | Complex of UBE2O with NAP1L1 and ubiquitylated uL2 | | Descriptor: | Nucleosome assembly protein 1-like 1, Ubiquitin,60S ribosomal protein L8,(E3-independent) E2 ubiquitin-conjugating enzyme fusion | | Authors: | Yip, M.C.J, Sedor, S.F, Shao, S. | | Deposit date: | 2022-04-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of client selection by the protein quality-control factor UBE2O.
Nat.Struct.Mol.Biol., 29, 2022
|
|
