4LXC
 
 | | The antimicrobial peptidase lysostaphin from Staphylococcus simulans | | Descriptor: | Lysostaphin, SULFATE ION, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
3R5Z
 
 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3OLJ
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) | | Descriptor: | Ribonucleoside-diphosphate reductase subunit M2, SODIUM ION | | Authors: | Chen, X.H, Xu, Z.J, Chen, B.E, Jiang, H.J, Yang, C.G, Zhu, W.L, Shao, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | hRRM2
To be Published
|
|
4LOS
 
 | | C1s CUB2-CCP1 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E, Gingras, A.R. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOT
 
 | | C1s CUB2-CCP1-CCP2 | | Descriptor: | Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L9H
 
 | | Crystal structure of the FP domain of human F-box protein Fbxo7(SeMet) | | Descriptor: | F-box only protein 7 | | Authors: | Du, Z, Huang, X, Shang, J, Yang, Y, Wang, G. | | Deposit date: | 2013-06-18 | | Release date: | 2014-01-15 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the FP domain of Fbxo7 reveals a novel mode of protein-protein interaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L9C
 
 | | Crystal structure of the FP domain of human F-box protein Fbxo7 (native) | | Descriptor: | F-box only protein 7, GLYCEROL | | Authors: | Du, Z, Huang, X, Shang, J, Yang, Y, Wang, G. | | Deposit date: | 2013-06-18 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the FP domain of Fbxo7 reveals a novel mode of protein-protein interaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OFS
 
 | | Crystal structure of a truncated catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrot, N.L, Marshall, J.J.T, Svergun, D.I, Crennell, S.J, Hough, D.W, van den Elsen, J.M.H, Danson, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex.
Biochem.J., 463, 2014
|
|
7JG4
 
 | | Human GAR transformylase in complex with GAR substrate and AGF131 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-(5-{3-[(1S,7R,8R,9S)-4-amino-2-oxo-7lambda~4~-thia-3,5-diazatetracyclo[4.3.0.0~1,7~.0~7,9~]nona-3,5-dien-8-yl]propyl}thiophene-2-carbonyl)-L-glutamic acid, SODIUM ION, ... | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG0
 
 | | Human GAR transformylase in complex with GAR substrate and AGF102 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG3
 
 | | Human GAR transformylase in complex with GAR substrate and AGF103 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]furan-2-carbonyl}-L-glutamic acid, SODIUM ION, ... | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
4I2T
 
 | |
4I2U
 
 | | Crystal structure of the reduced glutaredoxin from Chlorella sorokiniana T-89 in complex with glutathione | | Descriptor: | GLUTATHIONE, glutaredoxin | | Authors: | Nien, C.-Y, Cheng, C.-Y, Shaw, J.-F, Chen, Y.-T, Liu, J.-H. | | Deposit date: | 2012-11-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure and functional analysis of glutaredoxin from Chlorella sorokiniana T-89
To be Published
|
|
4OTX
 
 | | Structure of the anti-Francisella tularensis O-antigen antibody N203 Fab fragment | | Descriptor: | AZIDE ION, CHLORIDE ION, N203 heavy chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Yang, C.-Y, Madico, G, Perkins, H.M, Roche, M.I, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-02-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Characterization of Francisella tularensis O-Antigen Antibodies at the Low End of Antigen Reactivity.
Monoclon Antib Immunodiagn Immunother, 33, 2014
|
|
4PB0
 
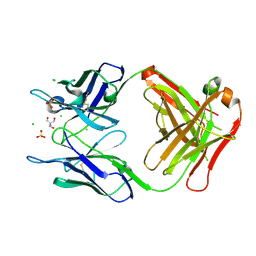 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab53 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ab53 heavy chain, Ab53 light chain, ... | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4QP5
 
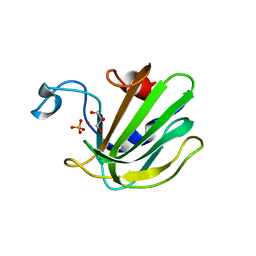 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate | | Descriptor: | GLYCEROL, Lysostaphin, PHOSPHATE ION, ... | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4PB9
 
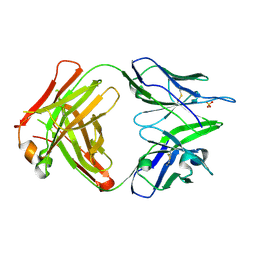 | | Structure of the Fab fragment of the anti-Francisella tularensis GroEL antibody Ab64 | | Descriptor: | Ab64 heavy chain, Ab64 light chain, SULFATE ION | | Authors: | Lu, Z, Rynkiewicz, M.J, Seaton, B.A, Sharon, J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | B-Cell Epitopes in GroEL of Francisella tularensis.
Plos One, 9, 2014
|
|
4QPB
 
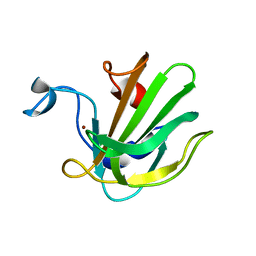 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate | | Descriptor: | 1,2-ETHANEDIOL, Lysostaphin, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4PMW
 
 | |
7K40
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | Authors: | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7JYC
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6D
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.48 A Resolution (Cryo-protected) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K3T
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-13 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7LJL
 
 | | Structure of the Enterobacter cloacae CD-NTase CdnD in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.W, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
