1FG5
 
 | | CRYSTAL STRUCTURE OF BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN. | | Descriptor: | N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE | | Authors: | Gastinel, L.N, Bignon, C, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-07-28 | | Release date: | 2001-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
6MRP
 
 | |
6D86
 
 | | Structure of the Bovine p85a BH domain | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, SULFATE ION | | Authors: | Moore, S.A, Marshall, J.D, Anderson, D.H. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-23 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Patient-derived mutations within the N-terminal domains of p85 alpha impact PTEN or Rab5 binding and regulation.
Sci Rep, 8, 2018
|
|
1G93
 
 | | CRYSTAL STRUCTURE OF THE BOVINE CATALYTIC DOMAIN OF ALPHA-1,3-GALACTOSYLTRANSFERASE IN THE PRESENCE OF UDP-GALACTOSE | | Descriptor: | MANGANESE (II) ION, MERCURY (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Gastinel, L.N, Bignon, C, Misra, A.K, Hindsgaul, O, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
1G8O
 
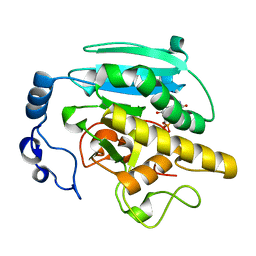 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NATIVE BOVINE ALPHA-1,3-GALACTOSYLTRANSFERASE CATALYTIC DOMAIN | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Gastinel, L.N, Bigon, C, Misra, A.K, Hindsgaul, O, Shaper, J.H, Joziasse, D.H. | | Deposit date: | 2000-11-20 | | Release date: | 2001-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases.
EMBO J., 20, 2001
|
|
2L9H
 
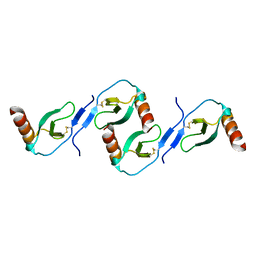 | | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data | | Descriptor: | C-C motif chemokine 5 | | Authors: | Wang, X, Watson, C.M, Sharp, J.S, Handel, T.M, Prestegard, J.H. | | Deposit date: | 2011-02-09 | | Release date: | 2011-06-22 | | Last modified: | 2011-08-24 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data.
Structure, 19, 2011
|
|
3ELQ
 
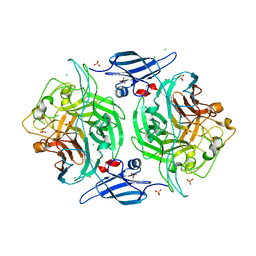 | | Crystal structure of a bacterial arylsulfate sulfotransferase | | Descriptor: | Arylsulfate sulfotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2A4Z
 
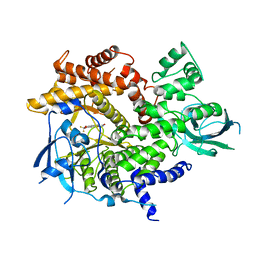 | | Crystal Structure of human PI3Kgamma complexed with AS604850 | | Descriptor: | (5E)-5-[(2,2-DIFLUORO-1,3-BENZODIOXOL-5-YL)METHYLENE]-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-06-30 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
3ETT
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase catalytic intermediate with 4-nitrophenol bound in the active site | | Descriptor: | Arylsulfate sulfotransferase, P-NITROPHENOL, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3ETS
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase catalytic intermediate with 4-methylumbelliferone bound in the active site | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Arylsulfate sulfotransferase, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2A5U
 
 | | Crystal Structure of human PI3Kgamma complexed with AS605240 | | Descriptor: | (5E)-5-(QUINOXALIN-6-YLMETHYLENE)-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
4PMW
 
 | |
3FW3
 
 | | Crystal Structure of soluble domain of CA4 in complex with Dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 4, SULFATE ION, ... | | Authors: | Greasley, S.E, Ferre, R.A.A, Paz, R, Wickersham, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Thioether benzenesulfonamide inhibitors of carbonic anhydrases II and IV: structure-based drug design, synthesis, and biological evaluation.
Bioorg.Med.Chem., 18, 2010
|
|
3GA4
 
 | | Crystal structure of Ost6L (photoreduced form) | | Descriptor: | 1,2-ETHANEDIOL, Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6, TETRAETHYLENE GLYCOL | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-16 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G9B
 
 | | Crystal structure of reduced Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G7Y
 
 | | Crystal structure of oxidized Ost6L | | Descriptor: | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit OST6 | | Authors: | Stirnimann, C.U, Grimshaw, J.P.A, Schulz, B.L, Brozzo, M.S, Fritsch, F, Glockshuber, R, Capitani, G, Gruetter, M.G, Aebi, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1RBQ
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Human GAR Tfase complex structure with polyglutamated
10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
1RBM
 
 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1RBY
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
7T63
 
 | |
1MEO
 
 | | human glycinamide ribonucleotide Transformylase at pH 4.2 | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase, SULFATE ION | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MEN
 
 | | complex structure of human GAR Tfase and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1RGR
 
 | | Cyclic Peptides Targeting PDZ Domains of PSD-95: Structural Basis for Enhanced Affinity and Enzymatic Stability | | Descriptor: | BETA-ALANINE, Presynaptic density protein 95, postsynaptic protein CRIPT peptide | | Authors: | Piserchio, A, Salinas, G.D, Li, T, Marshall, J, Spaller, M.R, Mierke, D.F. | | Deposit date: | 2003-11-12 | | Release date: | 2004-05-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Targeting Specific PDZ Domains of PSD-95; Structural Basis for Enhanced Affinity and Enzymatic Stability of a Cyclic Peptide.
Chem.Biol., 11, 2004
|
|
1MZY
 
 | | Crystal Structure of Nitrite Reductase | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MAGNESIUM ION | | Authors: | Guo, H, Olesen, K, Xue, Y, Shapliegh, J, Sjolin, L. | | Deposit date: | 2002-10-10 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The High resolution Crystal Structures of Nitrite Reductase and its mutant Met182Thr from Rhodobacter Sphaeroides Reveal a Gating Mechanism for the Electron Transfer to the Type 1 Copper Center
To be Published
|
|
1MEJ
 
 | | Human Glycinamide Ribonucleotide Transformylase domain at pH 8.5 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
