7WR4
 
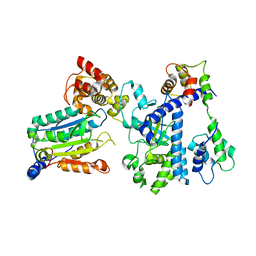 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
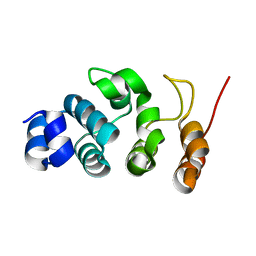 | |
7WR0
 
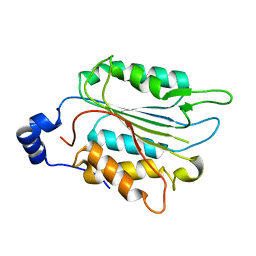 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
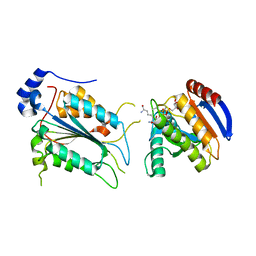 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3ZPQ
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-(piperazin-1- yl)-1H-indole bound (compound 19) | | Descriptor: | 4-(PIPERAZIN-1-YL)-1H-INDOLE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
3ZPR
 
 | | Thermostabilised turkey beta1 adrenergic receptor with 4-methyl-2-(piperazin-1-yl) quinoline bound | | Descriptor: | 4-METHYL-2-(PIPERAZIN-1-YL) QUINOLINE, BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Christopher, J.A, Congreve, M, Dore, A.S, Marshall, F.H, Myszka, D.G, Brown, J, Koglin, M, Tehan, B, Errey, J.C, Tate, C.G, Warne, T. | | Deposit date: | 2013-03-01 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biophysical Fragment Screening of the Beta1-Adrenergic Receptor: Identification of High Affinity Aryl Piperazine Leads Using Structure-Based Drug Design.
J.Med.Chem., 56, 2013
|
|
2B5A
 
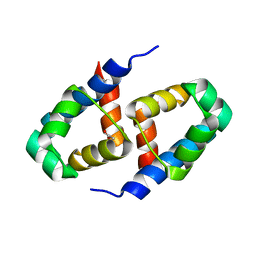 | | C.BclI, Control Element of the BclI Restriction-Modification System | | Descriptor: | ACETIC ACID, C.BclI | | Authors: | Sawaya, M.R, Zhu, Z, Mersha, F, Chan, S.H, Dabur, R, Xu, S.Y, Balendiran, G.K. | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Crystal Structure of the Restriction-Modification System Control Element C.BclI and Mapping of Its Binding Site.
Structure, 13, 2005
|
|
2C9E
 
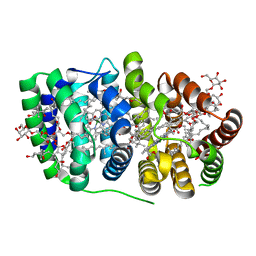 | | Peridinin-chlorophyll a protein, high-salt form | | Descriptor: | CHLOROPHYLL A, DIGALACTOSYL DIACYL GLYCEROL (DGDG), MAGNESIUM ION, ... | | Authors: | Schulte, T, Sharples, F.P, Hiller, R.G, Hofmann, E. | | Deposit date: | 2005-12-09 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structure of the High-Salt Form of the Peridinin-Chlorophyll A-Protein from the Dinoflagellate Amphidinium Carterae: Modulation of the Spectral Properties of Pigments by the Protein Environment.
Biochemistry, 48, 2009
|
|
5YXC
 
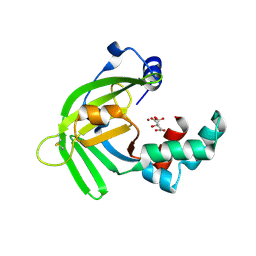 | | Crystal structure of Zinc binding protein ZinT in complex with citrate from E. coli | | Descriptor: | CITRIC ACID, Metal-binding protein ZinT, ZINC ION | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2017-12-04 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Biochem. Biophys. Res. Commun., 500, 2018
|
|
5Z7H
 
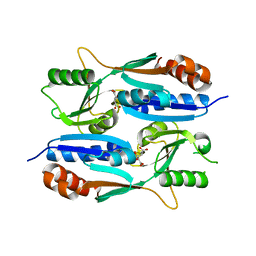 | |
5Z2C
 
 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
5ZZO
 
 | | Crystal structure of CcpE regulatory domain in complex with citrate from Staphyloccocus aureus | | Descriptor: | CITRATE ANION, LysR family transcriptional regulator | | Authors: | Chen, J, Wang, L, Shang, F, Xu, Y. | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Analysis of the Citrate-Responsive Mechanism of the Regulatory Domain of Catabolite Control Protein E from Staphylococcus aureus
Biochemistry, 57, 2018
|
|
5Y9Q
 
 | |
5Z72
 
 | |
3EIR
 
 | |
4JW1
 
 | | Crystal structure of N-terminal 618-residue fragment of LepB from Legionella pneumophila | | Descriptor: | CITRATE ANION, Effector protein B, GLYCEROL | | Authors: | Hu, L, Yao, Q, Zhu, Y, Shao, F. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-08 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural analyses of Legionella LepB reveal a new GAP fold that catalytically mimics eukaryotic RasGAP
Cell Res., 23, 2013
|
|
2P1W
 
 | |
4HCN
 
 | | Crystal structure of Burkholderia pseudomallei effector protein CHBP in complex with ubiquitin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Yao, Q, Cui, J, Zhu, Y, Shao, F. | | Deposit date: | 2012-09-30 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HCP
 
 | |
2Q8Y
 
 | |
3CVR
 
 | | Crystal structure of the full length IpaH3 | | Descriptor: | Invasion plasmid antigen | | Authors: | Zhu, Y, Shao, F. | | Deposit date: | 2008-04-19 | | Release date: | 2008-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Shigella effector reveals a new class of ubiquitin ligases
Nat.Struct.Mol.Biol., 15, 2008
|
|
1UKF
 
 | | Crystal Structure of Pseudomonas Avirulence Protein AvrPphB | | Descriptor: | Avirulence protein AVRPPH3 | | Authors: | Zhu, M, Shao, F, Innes, R.W, Dixon, J.E, Xu, Z. | | Deposit date: | 2003-08-21 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3EIT
 
 | |
3REY
 
 | | Thermostabilised adenosine A2A receptor in complex with XAC | | Descriptor: | Adenosine receptor A2a, N-(2-aminoethyl)-2-[4-(2,6-dioxo-1,3-dipropyl-2,3,6,7-tetrahydro-1H-purin-8-yl)phenoxy]acetamide | | Authors: | Dore, A.S, Robertson, N, Errey, J.C, Ng, I, Hollenstein, K, Tehan, B, Hurrell, E, Bennett, K, Congreve, M, Magnani, F, Tate, C.G, Weir, M, Marshall, F.H. | | Deposit date: | 2011-04-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.309 Å) | | Cite: | Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine
Structure, 19, 2011
|
|
3PWH
 
 | | Thermostabilised Adenosine A2A Receptor | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a | | Authors: | Dore, A.S, Robertson, N, Errey, J.C, Ng, I, Tehan, B, Hurrell, E, Magnani, F, Tate, C.G, Weir, M, Marshall, F.H. | | Deposit date: | 2010-12-08 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine
Structure, 19, 2011
|
|
