2GR1
 
 | |
6K4H
 
 | | Crystal structure of the PI5P4Kbeta-AMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
7EXZ
 
 | | DgpB-DgpC complex apo 2.5 angstrom | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-29 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7EXB
 
 | | DfgA-DfgB complex apo 2.4 angstrom | | Descriptor: | DfgB, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Mori, T, Senda, M, Senda, T, Abe, I. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
2GR2
 
 | |
6K4G
 
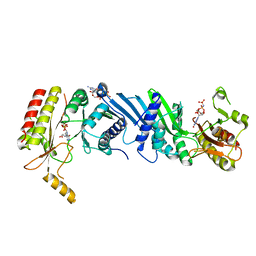 | | Crystal structure of the PI5P4Kbeta-GMPPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Senda, M, Senda, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 2022
|
|
2GQW
 
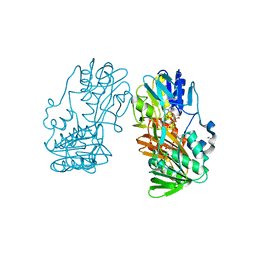 | | Crystal structure of Ferredoxin reductase, BphA4 (oxidized form) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2006-04-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2GR0
 
 | |
2YVF
 
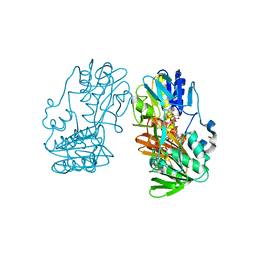 | | Crystal structure of ferredoxin reductase BPHA4 (hydroquinone) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, Ferredoxin reductase, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2YVG
 
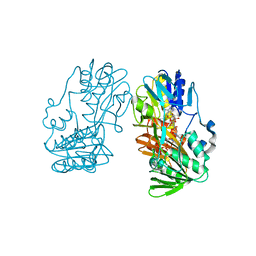 | | crystal structure of ferredoxin reductase, BPHA4 (blue-semiquinone) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2YVJ
 
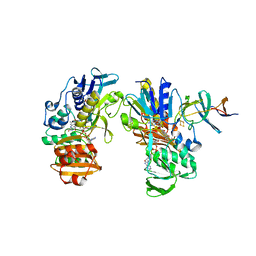 | | Crystal structure of the ferredoxin-ferredoxin reductase (BPHA3-BPHA4)complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
6ISV
 
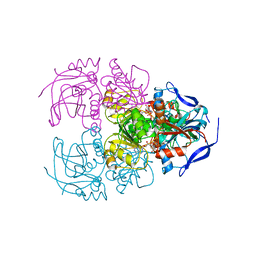 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | Descriptor: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
7EBT
 
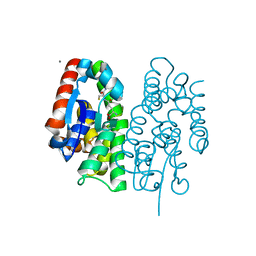 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in glutathione-bound form | | Descriptor: | CALCIUM ION, GLUTATHIONE, Glutathione transferase | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBW
 
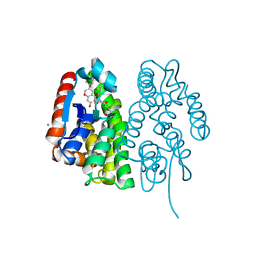 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in desmethylglycitein and glutathione-bound form | | Descriptor: | 6,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBU
 
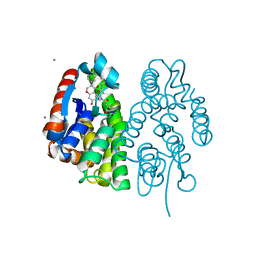 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in Daidzein- and glutathione-bound form | | Descriptor: | 7-hydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7E4D
 
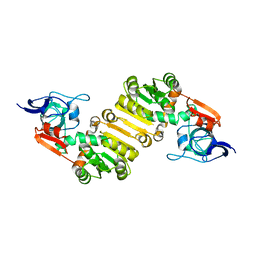 | | Crystal structure of PlDBR | | Descriptor: | Double Bond Reductase | | Authors: | Sugimoto, K, Senda, M, Senda, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
7EBV
 
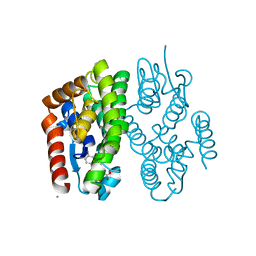 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in luteolin- and glutathione-bound form | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
4DVY
 
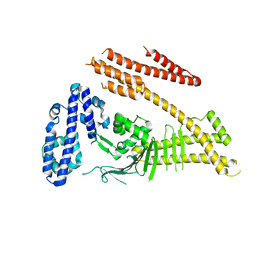 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
4DVZ
 
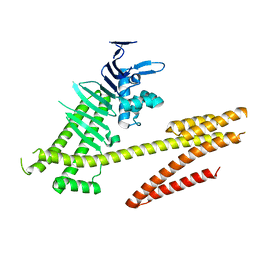 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
5XNY
 
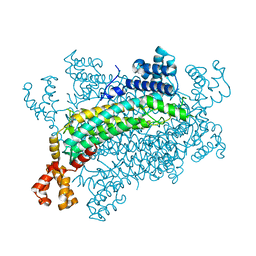 | | Crystal structure of CreD | | Descriptor: | CreD | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5XNZ
 
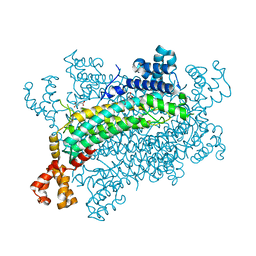 | | Crystal structure of CreD complex with fumarate | | Descriptor: | CreD, FUMARIC ACID | | Authors: | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | Deposit date: | 2017-05-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5XXP
 
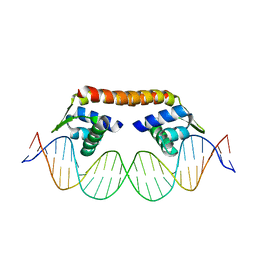 | | Crystal structure of CbnR_DBD-DNA complex | | Descriptor: | DNA (25-MER), LysR-type regulatory protein | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the DNA-binding domain of the LysR-type transcriptional regulator CbnR in complex with a DNA fragment of the recognition-binding site in the promoter region
FEBS J., 285, 2018
|
|
3WKU
 
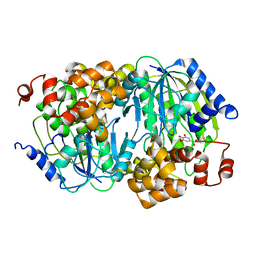 | | Crystal structure of the anaerobic DesB-gallate complex | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2013-10-31 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WR8
 
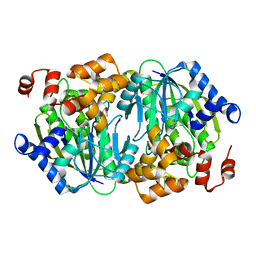 | | Crystal structure of DesB from Sphingobium sp. strain SYK-6 | | Descriptor: | FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
3WPM
 
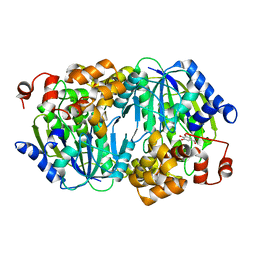 | | Crystal structure of the anaerobic DesB-gallate complex by co-crystallization | | Descriptor: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-01-14 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
