1JF7
 
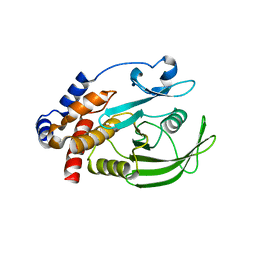 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
1LMQ
 
 | |
1T6E
 
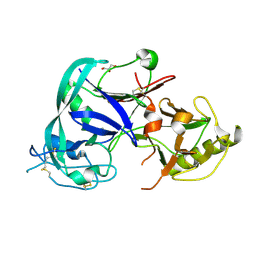 | | Crystal Structure of the Triticum aestivum xylanase inhibitor I | | Descriptor: | GLYCEROL, xylanase inhibitor | | Authors: | Sansen, S, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
J.Biol.Chem., 279, 2004
|
|
1LMO
 
 | |
3HD8
 
 | | Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase | | Descriptor: | Endo-1,4-beta-xylanase A, Xylanase inhibitor | | Authors: | Sansen, S, Pollet, A, Raedschelders, G, Gebruers, K, Rabijns, A, Courtin, C.M. | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA
Febs J., 276, 2009
|
|
2K69
 
 | |
2K68
 
 | |
2K67
 
 | |
1HLS
 
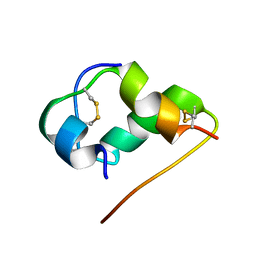 | | NMR STRUCTURE OF THE HUMAN INSULIN-HIS(B16) | | Descriptor: | INSULIN | | Authors: | Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1995-06-28 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of an engineered biologically potent insulin monomer, B16 Tyr-->His, as determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 33, 1994
|
|
1K8G
 
 | |
6RWI
 
 | | Fragment AZ-002 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-phenylthiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RWS
 
 | | Fragment AZ-009 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-6-(sulfanylmethyl)-1-benzothiophene-2-carboximidamide, Cellular tumor antigen p53, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RWH
 
 | | Fragment AZ-007 binding at a primary and secondary binding site of the the p53pT387/14-3-3 sigma complex | | Descriptor: | 14-3-3 protein sigma, 5-(1~{H}-imidazol-5-yl)-4-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
1LSP
 
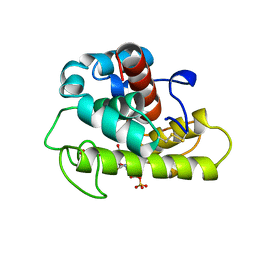 | | THE CRYSTAL STRUCTURE OF A BULGECIN-INHIBITED G-TYPE LYSOZYME FROM THE EGG-WHITE OF THE AUSTRALIAN BLACK SWAN. A COMPARISON OF THE BINDING OF BULGECIN TO THREE MURAMIDASES | | Descriptor: | BULGECIN A, LYSOZYME | | Authors: | Karlsen, S, Rao, Z.H, Hough, E, Isaacs, N.W. | | Deposit date: | 1995-02-09 | | Release date: | 1996-01-01 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of a bulgecin-inhibited g-type lysozyme from the egg white of the Australian black swan. A comparison of the binding of bulgecin to three muramidases.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6S9Q
 
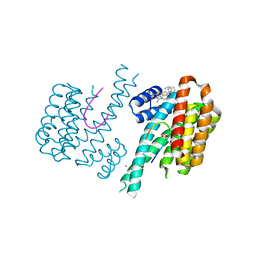 | | Fragment AZ-004 binding at a primary and secondary site in a p53pT387/14-3-3 complex | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-07-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
1A7F
 
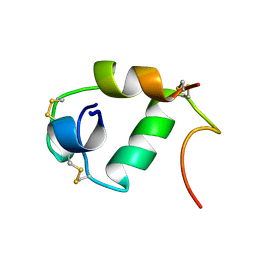 | |
6SIP
 
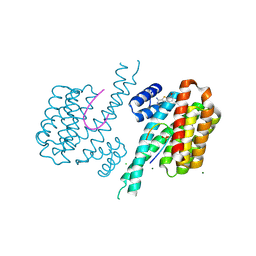 | | Fragment AZ-011 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-methoxy-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Leysen, S, Wolter, M, Guillory, X, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-08-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.600447 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6SIQ
 
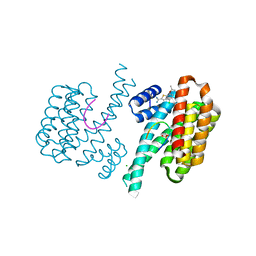 | | Fragment AZ-012 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-7-propan-2-yloxy-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Leysen, S, Wolter, M, Guillory, X, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-08-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60121 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6SIO
 
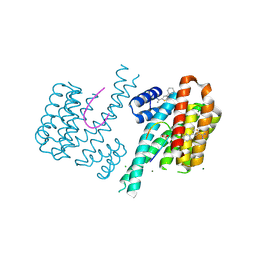 | | Fragment AZ-017 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-methyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Leysen, S, Wolter, M, Guillory, X, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-08-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60418 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RWU
 
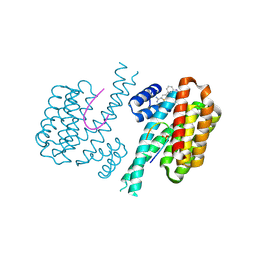 | | Fragment AZ-010 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, Cellular tumor antigen p53, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]propanamide | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6S40
 
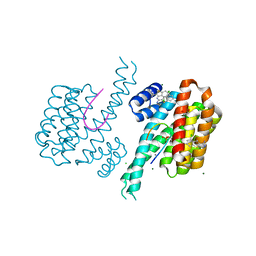 | | Fragment AZ-001 binding at the p53pT387/14-3-3 sigma interface and additional sites | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-1-benzothiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
1LMC
 
 | | THE CRYSTAL STRUCTURE OF A COMPLEX BETWEEN BULGECIN, A BACTERIAL METABOLITE, AND LYSOZYME FROM THE RAINBOW TROUT | | Descriptor: | BULGECIN A, LYSOZYME | | Authors: | Karlsen, S, Hough, E. | | Deposit date: | 1994-11-14 | | Release date: | 1996-01-01 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a complex between bulgecin, a bacterial metabolite, and lysozyme from the rainbow trout.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1Q1U
 
 | | Crystal structure of human FHF1b (FGF12b) | | Descriptor: | SULFATE ION, fibroblast growth factor homologous factor 1 | | Authors: | Olsen, S.K, Garbi, M, Zampieri, N, Eliseenkova, A.V, Ornitz, D.M, Goldfarb, M, Mohammadi, M. | | Deposit date: | 2003-07-22 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs
J.Biol.Chem., 278, 2003
|
|
2R4S
 
 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
2R4R
 
 | | Crystal structure of the human beta2 adrenoceptor | | Descriptor: | Beta-2 adrenergic receptor, antibody for beta2 adrenoceptor, heavy chain, ... | | Authors: | Rasmussen, S.G.F, Choi, H.J, Rosenbaum, D.M, Kobilka, T.S, Thian, F.S, Edwards, P.C, Burghammer, M, Ratnala, V.R, Sanishvili, R, Fischetti, R.F, Schertler, G.F, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human beta2 adrenergic G-protein-coupled receptor.
Nature, 450, 2007
|
|
