4WN3
 
 | | Crystal structure of Saccharomyces cerevisiae OMP synthase in complex with PRP(NH)P | | Descriptor: | MAGNESIUM ION, Orotate phosphoribosyltransferase 1, [[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]oxy-oxidanyl-phosphoryl]amino]phosphonic acid | | Authors: | Bang, M.B, Molich, U, Hansen, M.R, Grubmeyer, C, Harris, P. | | Deposit date: | 2014-10-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae OMP synthase in complex with PRP(NH)P
To Be Published
|
|
6WNL
 
 | | human Artemis/SNM1C catalytic domain, crystal form 2 | | Descriptor: | Protein artemis, ZINC ION | | Authors: | Karim, F, Liu, S, Laciak, A.R, Volk, L, Rosenblum, M, Curtis, R, Huang, N, Carr, G, Zhu, G. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural analysis of the catalytic domain of Artemis endonuclease/SNM1C reveals distinct structural features.
J.Biol.Chem., 295, 2020
|
|
6CQG
 
 | |
6NHK
 
 | | Mortalin nucleotide binding domain in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Page, R.C, Moseng, M.A, Nix, J.C. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.777 Å) | | Cite: | Biophysical Consequences of EVEN-PLUS Syndrome Mutations for the Function of Mortalin.
J.Phys.Chem.B, 123, 2019
|
|
3U8M
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3920 (1-(6-bromopyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
3U8K
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3573 (1-(5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
3U8N
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3950 (1-(6-bromo-5-ethoxypyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(6-bromo-5-ethoxypyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
7QOO
 
 | | Structure of the human inner kinetochore CCAN complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Vetter, I.R, Pesenti, M, Raisch, T. | | Deposit date: | 2021-12-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization.
Mol.Cell, 82, 2022
|
|
6XYA
 
 | | Cap-binding domain of SFTSV L protein | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, SODIUM ION | | Authors: | Gogrefe, N, Guenther, S, Rosenthal, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
6ZMV
 
 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZM8
 
 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
8ASG
 
 | | Structure of the SFTSV L protein bound in a resting state [RESTING] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*AP*UP*GP*A*)-3'), RNA (5'-R(*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U*)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8ASB
 
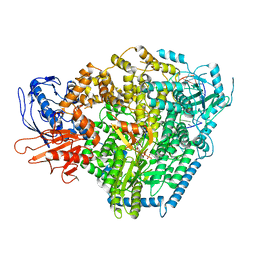 | | Structure of the SFTSV L protein stalled at early elongation with the endonuclease domain in a raised conformation [EARLY-ELONGATION-ENDO] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8ASD
 
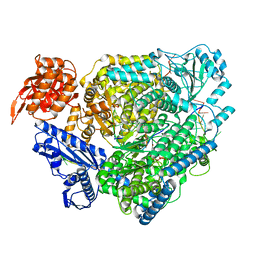 | | Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS7
 
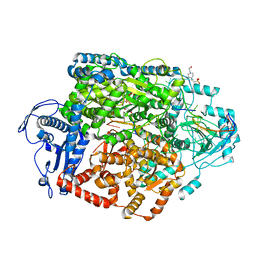 | | Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS6
 
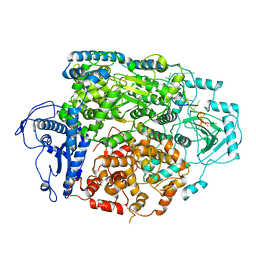 | | Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK] | | Descriptor: | RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*C)-3'), RNA-dependent RNA-polymerase L protein | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
5ML9
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GNF
 
 | | Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase, ... | | Authors: | Cuesta-Seijo, J.A, Nielsen, M.M, Palcic, M.M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of theCatalyticDomain ofArabidopsis thalianaStarch Synthase IV, of Granule Bound Starch Synthase From CLg1 and of Granule Bound Starch Synthase I ofCyanophora paradoxaIllustrate Substrate Recognition in Starch Synthases.
Front Plant Sci, 9, 2018
|
|
6H5Q
 
 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GNG
 
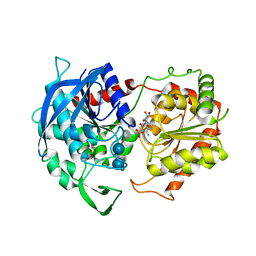 | | Granule Bound Starch Synthase I from Cyanophora paradoxa bound to acarbose and ADP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Granule-bound starch synthase | | Authors: | Cuesta-Seijo, J.A, Nielsen, M.M, Palcic, M.M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of theCatalyticDomain ofArabidopsis thalianaStarch Synthase IV, of Granule Bound Starch Synthase From CLg1 and of Granule Bound Starch Synthase I ofCyanophora paradoxaIllustrate Substrate Recognition in Starch Synthases.
Front Plant Sci, 9, 2018
|
|
6H5S
 
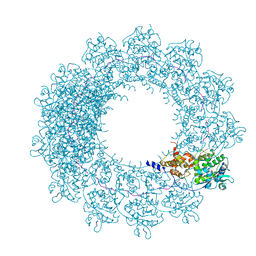 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5FYG
 
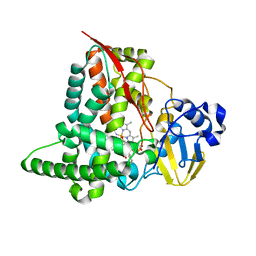 | | Structure of CYP153A from Marinobacter aquaeolei in complex with hydroxydodecanoic acid | | Descriptor: | 12-HYDROXYDODECANOIC ACID, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh-Azari, H.-R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
5GTU
 
 | |
6ZBA
 
 | | Crystal structure of PDE4D2 in complex with inhibitor LEO39652 | | Descriptor: | 1,2-ETHANEDIOL, 2-methylpropyl 1-[8-methoxy-5-(1-oxidanylidene-3~{H}-2-benzofuran-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]cyclopropane-1-carboxylate, DIMETHYL SULFOXIDE, ... | | Authors: | Akutsu, M, Hakansson, M, Welin, M, Svensson, A, Logan, D.T, Sorensen, M.D. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Early Clinical Development of Isobutyl 1-[8-Methoxy-5-(1-oxo-3 H -isobenzofuran-5-yl)-[1,2,4]triazolo[1,5- a ]pyridin-2-yl]cyclopropanecarboxylate (LEO 39652), a Novel "Dual-Soft" PDE4 Inhibitor for Topical Treatment of Atopic Dermatitis.
J.Med.Chem., 63, 2020
|
|
