1E8L
 
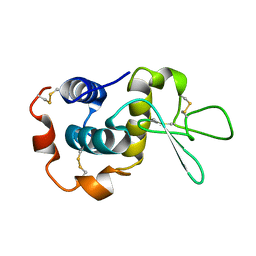 | | NMR solution structure of hen lysozyme | | Descriptor: | LYSOZYME | | Authors: | Schwalbe, H, Grimshaw, S.B, Spencer, A, Buck, M, Boyd, J, Dobson, C.M, Redfield, C, Smith, L.J. | | Deposit date: | 2000-09-27 | | Release date: | 2000-10-09 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | A refined solution structure of hen lysozyme determined using residual dipolar coupling data.
Protein Sci., 10, 2001
|
|
6XXB
 
 | |
6XXA
 
 | |
6XWW
 
 | |
6XWJ
 
 | | Constitutive decay element CDE2 from human 3'UTR | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*CP*UP*AP*AP*UP*AP*UP*UP*UP*AP*GP*GP*CP*AP*CP*C)-3') | | Authors: | Schwalbe, H, Binas, O. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
9EOW
 
 | |
6TQB
 
 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE1 SL RNA motif | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
4W9A
 
 | | Crystal structure of Gamma-B Crystallin expressed in E. coli based on mRNA variant 2 | | Descriptor: | Gamma-crystallin B | | Authors: | Kudlinzki, D, Buhr, F, Linhard, V.L, Jha, S, Komar, A.A, Schwalbe, H. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Two synonymous gene variants encode proteins with identical sequence, but different folding conformations.
To Be Published
|
|
4J23
 
 | | Low resolution crystal structure of the FGFR2D2D3/FGF1/SR128545 complex | | Descriptor: | Fibroblast growth factor 1, Fibroblast growth factor receptor 2 | | Authors: | Kudlinzki, D, Saxena, K, Sreeramulu, S, Schieborr, U, Dreyer, M, Schreuder, H, Schwalbe, H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.882 Å) | | Cite: | Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling.
Cancer Cell, 23, 2013
|
|
2W0G
 
 | |
6TQA
 
 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE2 SL RNA motif | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA (5'-R(P*GP*GP*UP*GP*CP*CP*UP*AP*AP*UP*AP*UP*UP*UP*AP*GP*GP*CP*AP*CP*(CCC))-3'), ... | | Authors: | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
5W72
 
 | |
8Q5B
 
 | |
7CLV
 
 | | Solution structure of mitochondrial Tim23 channel in complex with a signaling peptide | | Descriptor: | COX4 isoform 1, TIM23 isoform 1 | | Authors: | Zhou, S, Ruan, M.S, Li, Y.Y, Yang, J, Richter, C, Schwalbe, H, Shen, B, Wang, J.F. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the voltage-gated Tim23 channel in complex with a mitochondrial presequence peptide.
Cell Res., 31, 2021
|
|
1CFF
 
 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
7A05
 
 | | NMR structure of D3-D4 domains of Vibrio vulnificus ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Qureshi, N.S, Matzel, T, Cetiner, E.C, Schnieders, S, Jonker, H.R.A, Schwalbe, H, Fuertig, B. | | Deposit date: | 2020-08-06 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Vibrio vulnificus ribosomal protein S1 domains D3 and D4 provides insights into molecular recognition of single-stranded RNAs.
Nucleic Acids Res., 49, 2021
|
|
8PH4
 
 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
7QG7
 
 | | SARS-CoV-2 macrodomain Nsp3b bound to the remdesivir nucleoside GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Wollenhaupt, J, Linhard, V, Sreeramulu, S, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
4W9B
 
 | | Crystal structure of Gamma-B Crystallin expressed in E. coli based on mRNA variant 1 | | Descriptor: | Gamma-crystallin B | | Authors: | Kudlinzki, D, Buhr, F, Linhard, V.L, Jha, S, Komar, A.A, Schwalbe, H. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | Two synonymous gene variants encode proteins with identical sequence, but different folding conformations.
To Be Published
|
|
4WIH
 
 | | Crystal structure of cAMP-dependent Protein Kinase A from Cricetulus griseus | | Descriptor: | cAMP Dependent Protein Kinase Inhibitor PKI-tide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Kudlinzki, D, Linhard, V.L, Saxena, K, Dreyer, M, Schwalbe, H. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | High-resolution crystal structure of cAMP-dependent protein kinase from Cricetulus griseus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4YMH
 
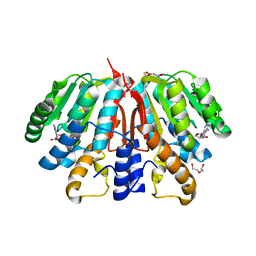 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
3POK
 
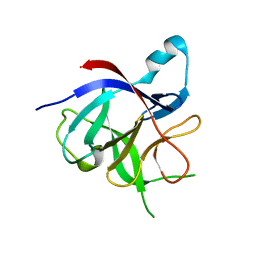 | | Interleukin-1-beta LBT L3 Mutant | | Descriptor: | Interleukin-1 beta | | Authors: | Barthelmes, K, Reynolds, A.M, Peisach, E, Jonker, H.R.A, DeNunzio, N.J, Allen, K.N, Imperiali, B, Schwalbe, H. | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering encodable lanthanide-binding tags into loop regions of proteins.
J.Am.Chem.Soc., 133, 2011
|
|
4YMG
 
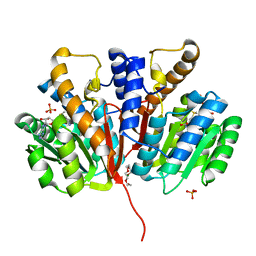 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
3RKF
 
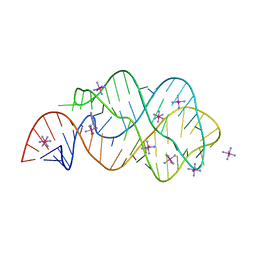 | | Crystal structure of guanine riboswitch C61U/G37A double mutant bound to thio-guanine | | Descriptor: | 2-amino-1,9-dihydro-6H-purine-6-thione, COBALT HEXAMMINE(III), Guanine riboswitch | | Authors: | Buck, J, Wacker, A, Warkentin, E, Woehnert, J, Wirmer-Bartoschek, J, Schwalbe, H. | | Deposit date: | 2011-04-18 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Influence of ground-state structure and Mg2+ binding on folding kinetics of the guanine-sensing riboswitch aptamer domain.
Nucleic Acids Res., 39, 2011
|
|
4QE8
 
 | | FXR with DM175 and NCoA-2 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 4-({2-[(4-tert-butylbenzoyl)amino]benzoyl}amino)benzoic acid, ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Sreeramulu, S, Nilsson, E, Dekker, N, Wissler, L, Bamberg, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | FXR with DM175 and NCoA-2 peptide
To be Published
|
|
