6BZD
 
 | |
6DXO
 
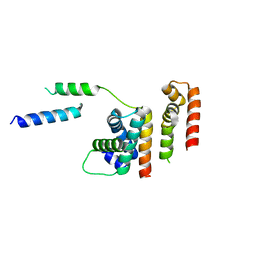 | | 1.8 A structure of RsbN-BldN complex. | | Descriptor: | BldN, RNA polymerase ECF-subfamily sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Nucleic Acids Res., 46, 2018
|
|
1YM8
 
 | |
7TEA
 
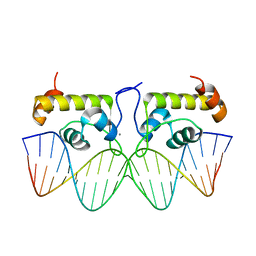 | | Crystal structure of S. aureus GlnR-DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*AP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*GP*TP*CP*AP*GP*AP*TP*TP*AP*TP*CP*TP*GP*AP*CP*AP*CP*G)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-01-04 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
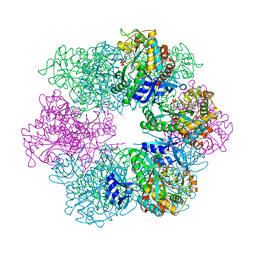
7TDP
 
 | |
7TEC
 
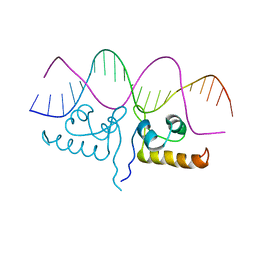 | |
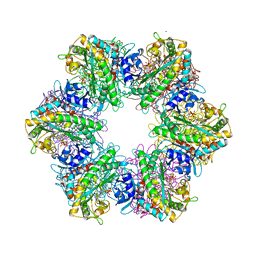
7TDV
 
 | |
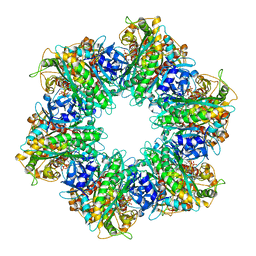
7TEN
 
 | |
7U3D
 
 | | Structure of S. venezuelae GlgX-c-di-GMP-acarbose complex (4.6) | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7U3B
 
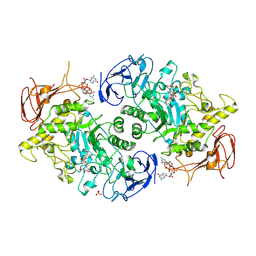 | | Structure of S. venezuelae GlgX bound to c-di-GMP and acarbose (pH 8.5) | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX, ... | | Authors: | Schumacher, M.A, Tschowri, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7U39
 
 | |
7U02
 
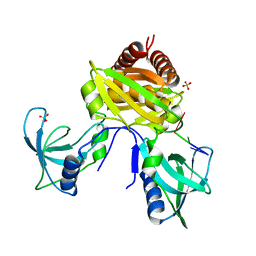 | | Structure of the C. crescentus DriD C-domain bound to ssDNA | | Descriptor: | DNA (5'-D(P*AP*CP*G)-3'), SULFATE ION, WYL domain-containing protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
7TZV
 
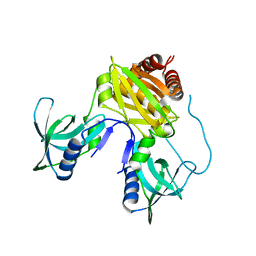 | | Structure of DriD C-domain bound to 9mer ssDNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | Authors: | Schumacher, M.A, Laub, M. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
7U3A
 
 | | Structure of the Streptomyces venezuelae GlgX-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
1PNR
 
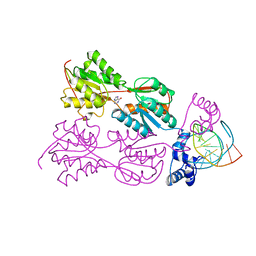 | | PURINE REPRESSOR-HYPOXANTHINE-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1995-03-29 | | Release date: | 1995-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
1DBQ
 
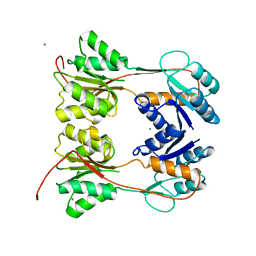 | | DNA-BINDING REGULATORY PROTEIN | | Descriptor: | MAGNESIUM ION, PURINE REPRESSOR | | Authors: | Schumacher, M.A, Choi, K.Y, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-02-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of corepressor-mediated specific DNA binding by the purine repressor.
Cell(Cambridge,Mass.), 83, 1995
|
|
1SDK
 
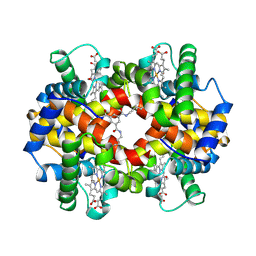 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
1RZR
 
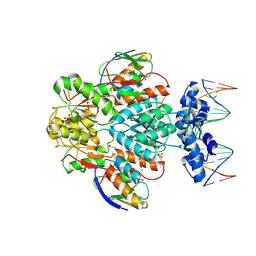 | | crystal structure of transcriptional regulator-phosphoprotein-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Glucose-resistance amylase regulator, ... | | Authors: | Schumacher, M.A, Allen, G.S, Brennan, R.G. | | Deposit date: | 2003-12-27 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXI
 
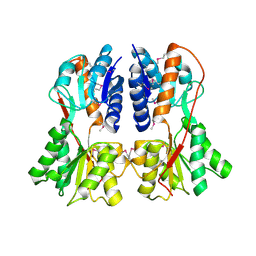 | | Structure of apo transcription regulator B. megaterium | | Descriptor: | Glucose-resistance amylase regulator, MAGNESIUM ION | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXG
 
 | | Structural studies on the apo transcription factor form B. megaterium | | Descriptor: | 2-PHENYLAMINO-ETHANESULFONIC ACID, Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXH
 
 | | apo structure of B. megaterium transcription regulator | | Descriptor: | Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1UPF
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V BOUND TO THE DRUG 5-FLUOROURACIL | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1UPU
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1SDL
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
5I41
 
 | | Structure of the apo RacA DNA binding domain | | Descriptor: | Chromosome-anchoring protein RacA | | Authors: | schumacher, M.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
