8EBL
 
 | |
8QBN
 
 | |
4YII
 
 | |
8C07
 
 | |
8QE8
 
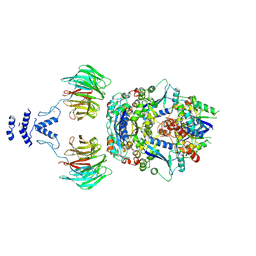 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | 著者 | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | 登録日 | 2023-08-30 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
6WY6
 
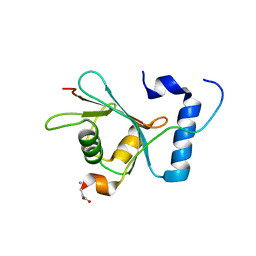 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | 分子名称: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | 著者 | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | 登録日 | 2020-05-12 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
7Q51
 
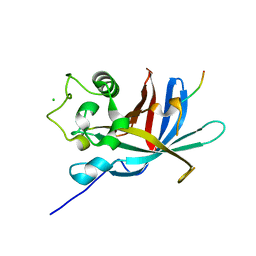 | | yeast Gid10 bound to a Phe/N-peptide | | 分子名称: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7Q50
 
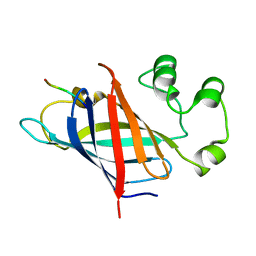 | | human Gid4 bound to a Phe/N-peptide | | 分子名称: | FDVSWFMG peptide, Glucose-induced degradation protein 4 homolog | | 著者 | Chrustowicz, J, Sherpa, D, Loke, M.S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
7QQY
 
 | |
7Q4Y
 
 | |
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | 分子名称: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-10-27 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
8Q7E
 
 | |
8Q7H
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | 分子名称: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | 著者 | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7Z8R
 
 | | CAND1-CUL1-RBX1 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Schulman, B.A. | | 登録日 | 2022-03-18 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
1Y8X
 
 | | Structural basis for recruitment of Ubc12 by an E2-binding domain in NEDD8's E1 | | 分子名称: | Ubiquitin-activating enzyme E1C, Ubiquitin-conjugating enzyme E2 M | | 著者 | Huang, D.T, Paydar, A, Zhuang, M, Waddell, M.B, Holton, J.M, Schulman, B.A. | | 登録日 | 2004-12-13 | | 公開日 | 2005-02-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1.
Mol.Cell, 17, 2005
|
|
7ZBZ
 
 | |
7Z8V
 
 | |
7Z8T
 
 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Schulman, B.A. | | 登録日 | 2022-03-18 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7ZBW
 
 | | CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Schulman, B.A. | | 登録日 | 2022-03-24 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8EBN
 
 | |
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | 分子名称: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-10-27 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
9BC9
 
 | | Structure of KLHDC2 bound to SJ10278 | | 分子名称: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[2-chloro-4-(methoxymethoxy)phenyl]-1,3-thiazol-4-yl}acetyl)glycine | | 著者 | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | 登録日 | 2024-04-08 | | 公開日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
9BCC
 
 | | Structure of KLHDC2 bound to SJ46418 | | 分子名称: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-({2-[8-(2-methoxyethoxy)naphthalen-2-yl]-1,3-thiazol-4-yl}acetyl)glycine | | 著者 | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | 登録日 | 2024-04-08 | | 公開日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
8RHZ
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | 分子名称: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | 著者 | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2023-12-17 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9BCA
 
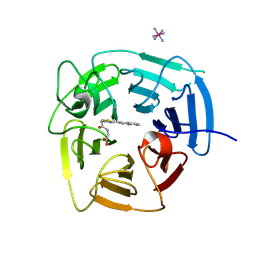 | | Structure of KLHDC2 bound to SJ46411 | | 分子名称: | COBALT HEXAMMINE(III), Kelch domain-containing protein 2, N-{[2-(naphthalen-2-yl)-1,3-thiazol-4-yl]acetyl}glycine | | 著者 | Scott, D.C, Dharuman, S, Griffith, E, Chai, S.C, Ronnebaum, J, King, M.T, Tangallapally, R, Lee, C, Gee, C.T, Lee, H.W, Ochoada, J, Miller, D.J, Jayasinghe, T, Paulo, J.A, Elledge, S.J, Harper, J.W, Chen, T, Lee, R.E, Schulman, B.A. | | 登録日 | 2024-04-08 | | 公開日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Principles of isoform-specific targeted protein degradation engaging the C-degron E3 KLHDC2
To Be Published
|
|
