1JT0
 
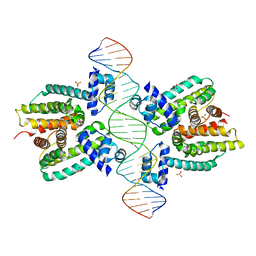 | | Crystal structure of a cooperative QacR-DNA complex | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, QACA operator, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-20 | | Release date: | 2002-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR.
EMBO J., 21, 2002
|
|
1JTX
 
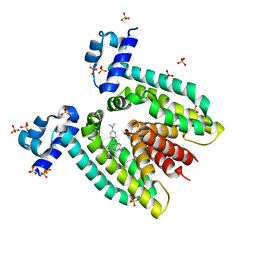 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to crystal violet | | Descriptor: | CRYSTAL VIOLET, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JT6
 
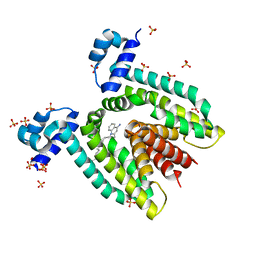 | | Crystal structure of the multidrug binding protein QacR bound to dequalinium | | Descriptor: | DEQUALINIUM, Hypothetical transcriptional regulator IN QACA 5'region, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1Q89
 
 | |
1JUS
 
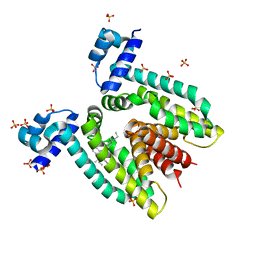 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to rhodamine 6G | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, RHODAMINE 6G, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-27 | | Release date: | 2001-12-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JTY
 
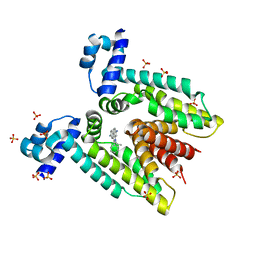 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to ethidium | | Descriptor: | ETHIDIUM, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JUM
 
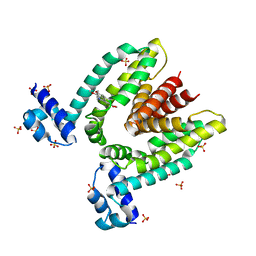 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to the natural drug berberine | | Descriptor: | BERBERINE, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1DH3
 
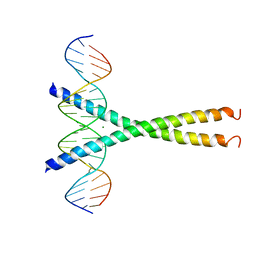 | |
5U1J
 
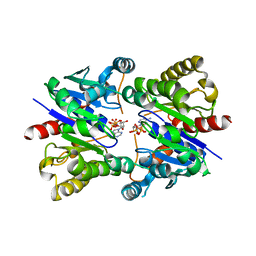 | | Structure of pNOB8 ParA bound to nonspecific DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*TP*AP*AP*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*AP*TP*GP*AP*CP*AP*CP*G)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation.
Genes Dev., 31, 2017
|
|
1JLS
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE URACIL/CPR 2 MUTANT C128V | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ullman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JLR
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE GTP COMPLEX 2 MUTANT C128V | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Uracil Phosphoribosyltransferase | | Authors: | Schumacher, M.A, Bashor, C.J, Otsu, K, Zu, S, Parry, R, Ulmman, B, Brennan, R.G. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1J4X
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE C124S MUTANT-PEPTIDE COMPLEX | | Descriptor: | DDE(AHP)(TPO)G(PTR)VATR, DUAL SPECIFICITY PROTEIN PHOSPHATASE 3 | | Authors: | Schumacher, M.A, Todd, J.L, Tanner, K.G, Denu, J.M. | | Deposit date: | 2001-12-13 | | Release date: | 2001-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein Phosphatase.
Biochemistry, 41, 2002
|
|
1KQ1
 
 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | Descriptor: | ACETIC ACID, Host Factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1UNC
 
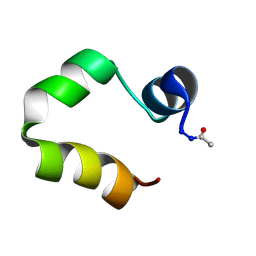 | | Solution structure of the human villin C-terminal headpiece subdomain | | Descriptor: | VILLIN 1 | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
4V15
 
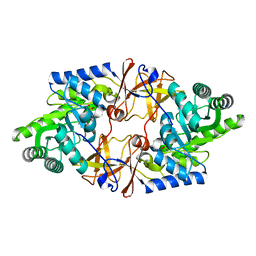 | | Crystal structure of D-threonine aldolase from Alcaligenes xylosoxidans | | Descriptor: | D-THREONINE ALDOLASE, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Uhl, M.K, Oberdorfer, G, Steinkellner, G, Riegler, L, Schuermann, M, Gruber, K. | | Deposit date: | 2014-09-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of D-Threonine Aldolase from Alcaligenes Xylosoxidans Provides Insight Into a Metal Ion Assisted Plp-Dependent Mechanism.
Plos One, 10, 2015
|
|
5HW8
 
 | |
2PUG
 
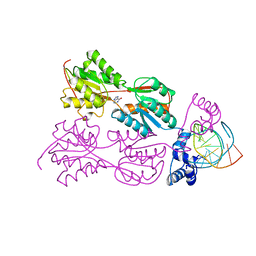 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
2PUF
 
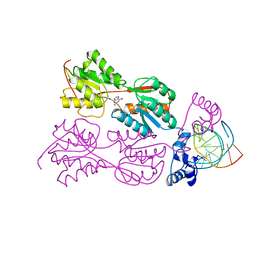 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Lu, F, Schumacher, M.A, Arvidson, D.N, Haldimann, A, Wanner, B.L, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190.
Biochemistry, 37, 1998
|
|
5HW6
 
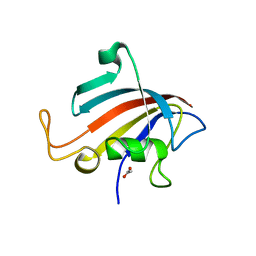 | |
4Z58
 
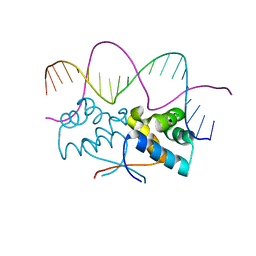 | | HipB-O3 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
5HW7
 
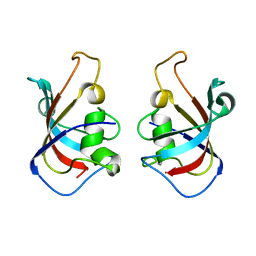 | |
1A3H
 
 | |
5HWC
 
 | |
7QUY
 
 | | Alcohol Dehydrogenase from Thauera aromatica complexed with NADH | | Descriptor: | 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
4RMI
 
 | | Human Sirt2 in complex with SirReal1 and Ac-Lys-OTC peptide | | Descriptor: | Ac-Lys-OTC peptide, N-(5-benzyl-1,3-thiazol-2-yl)-2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]acetamide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
