4Z5H
 
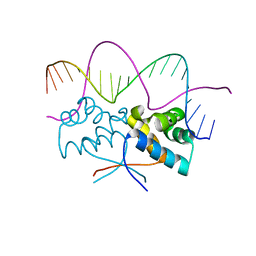 | | HipB(S29A)-O2 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*TP*CP*AP*CP*TP*AP*AP*AP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z59
 
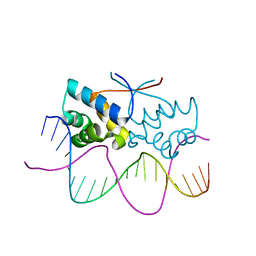 | | HipB-O4 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
2A3H
 
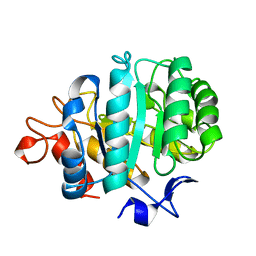 | | CELLOBIOSE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHERANS AT 2.0 A RESOLUTION | | Descriptor: | ENDOGLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Davies, G.J, Brzozowski, A.M, Andersen, K, Schulein, M. | | Deposit date: | 1998-01-22 | | Release date: | 1999-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Bacillus agaradherans family 5 endoglucanase at 1.6 A and its cellobiose complex at 2.0 A resolution
Biochemistry, 37, 1998
|
|
3GXQ
 
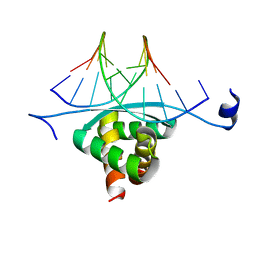 | | Structure of ArtA and DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*AP*CP*AP*TP*G)-3'), DNA (5'-D(*AP*CP*AP*TP*GP*TP*CP*AP*TP*GP*T)-3'), Putative regulator of transfer genes ArtA | | Authors: | Ni, L, Firth, N, Schumacher, M.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes.
Nucleic Acids Res., 37, 2009
|
|
4BV0
 
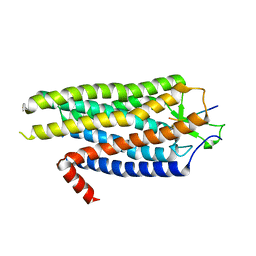 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4PYD
 
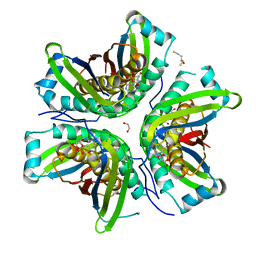 | | MoaC in complex with cPMP crystallized in space group P212121 | | Descriptor: | (2R,4AR,5AR,11AR,12AS)-8-AMINO-2-HYDROXY-4A,5A,9,11,11A,12A-HEXAHYDRO[1,3,2]DIOXAPHOSPHININO[4',5':5,6]PYRANO[3,2-G]PTERIDINE-10,12(4H,6H)-DIONE 2-OXIDE, 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Tonthat, N.K, Hover, B.M, Yokoyama, K, Schumacher, M.A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4B7M
 
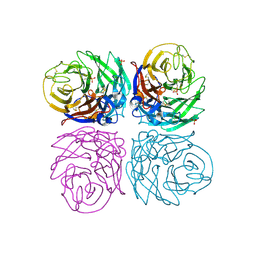 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
4B7Q
 
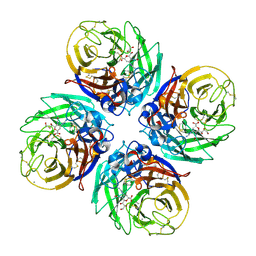 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
3JV1
 
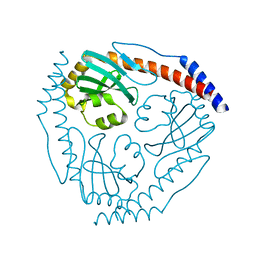 | |
4B7R
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
6HL3
 
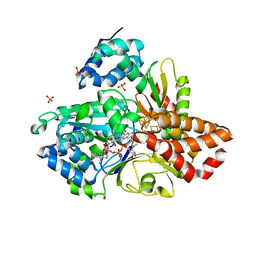 | | wild-type NuoEF from Aquifex aeolicus - oxidized form bound to NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6HL2
 
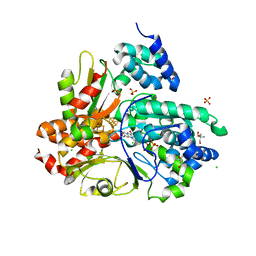 | | wild-type NuoEF from Aquifex aeolicus - oxidized form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gerhardt, S, Friedrich, T, Einsle, O, Gnandt, E, Schulte, M, Fiegen, D. | | Deposit date: | 2018-09-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
1QQB
 
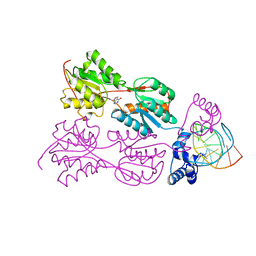 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*TP*CP*GP*AP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
1QP4
 
 | | PURINE REPRESSOR-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*TP*CP*GP*AP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
417D
 
 | | A THYMINE-LIKE BASE ANALOGUE FORMS WOBBLE PAIRS WITH ADENINE | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*(C46)P*G)-3') | | Authors: | Lin, P.K.T, Schuerman, M.H, Moore, G.S, Van Meervelt, L, Loakes, D, Brown, D.M, Moore, M.H. | | Deposit date: | 1998-07-15 | | Release date: | 1998-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A thymine-like base analogue forms wobble pairs with adenine in a Z-DNA duplex.
J.Mol.Biol., 282, 1998
|
|
1QP0
 
 | | PURINE REPRESSOR-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*CP*CP*GP*GP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
1QP7
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*CP*CP*GP*GP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
1QPZ
 
 | | PURINE REPRESSOR-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
1QQA
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*GP*CP*GP*CP*TP*TP*GP*CP*GP*T)-3'), HYPOXANTHINE, PROTEIN (PURINE NUCLEOTIDE SYNTHESIS REPRESSOR) | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
4ENG
 
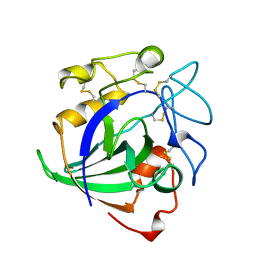 | | STRUCTURE OF ENDOGLUCANASE V CELLOHEXAOSE COMPLEX | | Descriptor: | ENDOGLUCANASE V CELLOHEXAOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
5HW8
 
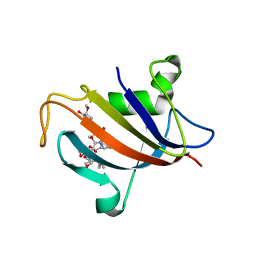 | |
2K33
 
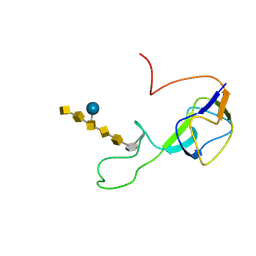 | | Solution structure of an N-glycosylated protein using in vitro glycosylation | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-[beta-D-glucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, AcrA | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F.H.-T. | | Deposit date: | 2008-04-18 | | Release date: | 2009-02-03 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
4PYA
 
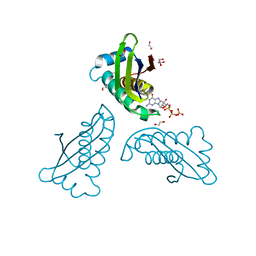 | | MoaC K51A in complex with 3',8-cH2GTP | | Descriptor: | (8S)-3',8-cyclo-7,8-dihydroguanosine 5'-triphosphate, 1,2-ETHANEDIOL, Molybdenum cofactor biosynthesis protein MoaC | | Authors: | Tonthat, N.K, Hover, B.M, Yokoyama, K, Schumacher, M.A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Mechanism of pyranopterin ring formation in molybdenum cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MPW
 
 | | Solution structure of the LysM region of the E. coli Intimin periplasmic domain | | Descriptor: | Intimin | | Authors: | Coles, M, Chaubey, M, Leo, J.C, Linke, D, Schuetz, M.C, Goetz, F, Autenrieth, I.B. | | Deposit date: | 2014-06-05 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan.
Mol.Microbiol., 95, 2015
|
|
4B7J
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-20 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
