4KOA
 
 | | Crystal Structure Analysis of 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti | | Descriptor: | 1,5-anhydro-D-fructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schu, M, Faust, A, Stosik, B, Kohring, G.-W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2013-05-11 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structure of substrate-free 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti 1021 reveals an open enzyme conformation.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3VEA
 
 | | Crystal Structure of matP-matS23mer | | Descriptor: | 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', 5'-D(*AP*GP*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*AP*CP*T)-3', Macrodomain Ter protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-01-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
3VEB
 
 | | Crystal Structure of Matp-matS | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*G)-3', 5'-D(*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*G)-3', CALCIUM ION, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-01-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
1J4X
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE C124S MUTANT-PEPTIDE COMPLEX | | Descriptor: | DDE(AHP)(TPO)G(PTR)VATR, DUAL SPECIFICITY PROTEIN PHOSPHATASE 3 | | Authors: | Schumacher, M.A, Todd, J.L, Tanner, K.G, Denu, J.M. | | Deposit date: | 2001-12-13 | | Release date: | 2001-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein Phosphatase.
Biochemistry, 41, 2002
|
|
6NOO
 
 | | Structure of Cyanothece McdA-AMPPNP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maintenance of carboxysome positioning A protein, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.
Nucleic Acids Res., 47, 2019
|
|
6UEP
 
 | |
6NOP
 
 | | Structure of Cyanothece McdA(D38A)-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobyrinic acid ac-diamide synthase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.
Nucleic Acids Res., 47, 2019
|
|
6GY1
 
 | | rat COMT in complex with inhibitor | | Descriptor: | 7-fluoranyl-5-(4-methylphenyl)sulfonyl-quinolin-8-ol, Catechol O-methyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of 8-Hydroxyquinolines as Inhibitors of Catechol O-Methyltransferase.
J. Med. Chem., 61, 2018
|
|
1WET
 
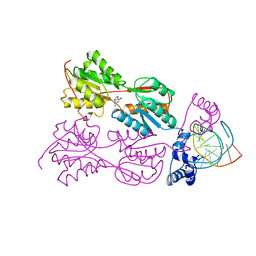 | | STRUCTURE OF THE PURR-GUANINE-PURF OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Glasfeld, A, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-04-27 | | Release date: | 1997-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity.
J.Biol.Chem., 272, 1997
|
|
3MKW
 
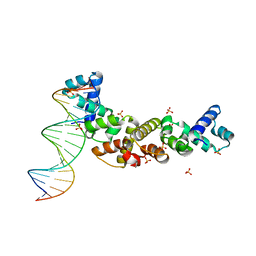 | | Structure of sopB(155-272)-18mer complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
2GID
 
 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
3M9A
 
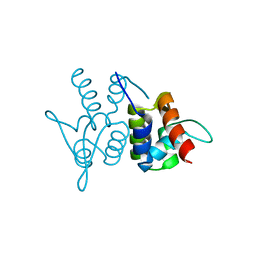 | | Protein structure of type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4GFK
 
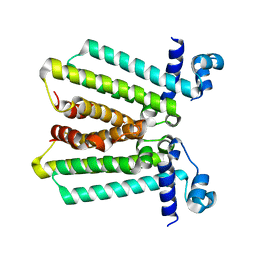 | | structures of NO factors | | Descriptor: | Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-08-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4W4G
 
 | | Postcleavage state of 70S bound to HigB toxin and AAA (lysine) codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2G66
 
 | |
2GIA
 
 | | Crystal structures of trypanosoma bruciei MRP1/MRP2 | | Descriptor: | ACETIC ACID, mitochondrial RNA-binding protein 1, mitochondrial RNA-binding protein 2 | | Authors: | Schumacher, M.A, Karamooz, E, Zikova, A, Trantirek, L, Lukes, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of T. brucei MRP1/MRP2 Guide-RNA Binding Complex Reveal RNA Matchmaking Mechanism.
Cell(Cambridge,Mass.), 126, 2006
|
|
9BE2
 
 | |
3BTI
 
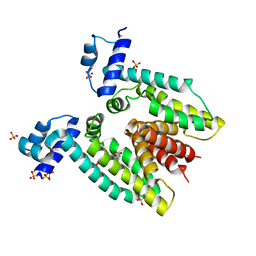 | |
3BTC
 
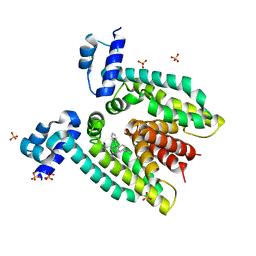 | |
3BTJ
 
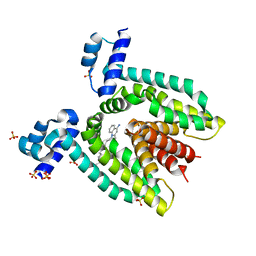 | |
3BTL
 
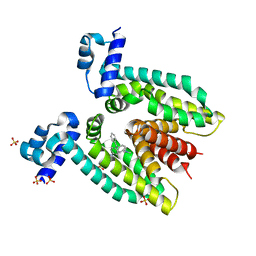 | |
6PFJ
 
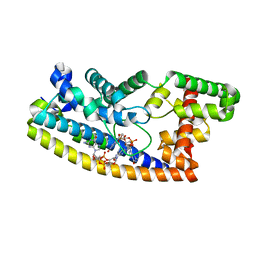 | | Structure of S. venezuelae RsiG-WhiG-(ci-di-GMP) complex, P64 crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-21 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6PFV
 
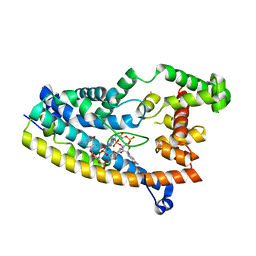 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
8SVD
 
 | |
6UEO
 
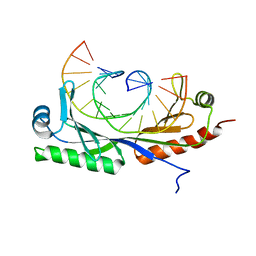 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
