8C5K
 
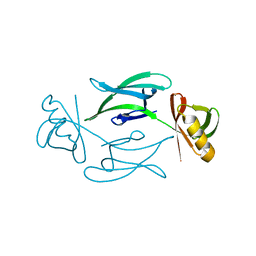 | | HEX-1 (in cellulo, in situ) crystallized and diffracted in High Five cells. Growth and SX data collection at 296 K on CrystalDirect plates | | Descriptor: | Woronin body major protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C51
 
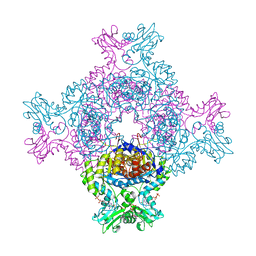 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD4
 
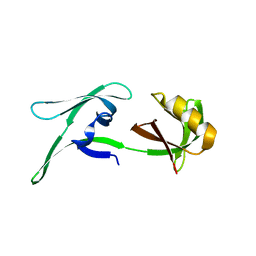 | | structure of HEX-1 from N. crassa crystallized in cellulo (cytosol), diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8CD6
 
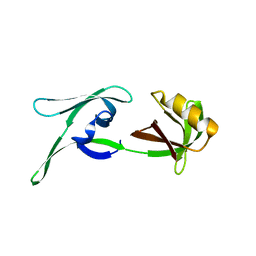 | | structure of HEX-1 (cyto V2) from N. crassa grown in living insect cells, diffracted at 100K and resolved using CrystFEL | | Descriptor: | Woronin body major protein | | Authors: | Boger, J, Schoenherr, R, Lahey-Rudolph, J.M, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | Deposit date: | 2023-01-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
1T40
 
 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 at ph 5 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | Authors: | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1F8G
 
 | | THE X-RAY STRUCTURE OF NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE FROM RHODOSPIRILLUM RUBRUM COMPLEXED WITH NAD+ | | Descriptor: | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Buckley, P.A, Baz Jackson, J, Schneider, T, White, S.A, Rice, D.W, Baker, P.J. | | Deposit date: | 2000-06-30 | | Release date: | 2001-06-30 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex.
Structure Fold.Des., 8, 2000
|
|
1T41
 
 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | Authors: | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3T8E
 
 | | Crystal structure of CerJ from Streptomyces tendae soaked with CerviK | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-08-01 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T5Y
 
 | | Crystal structure of CerJ from Streptomyces tendae - malonic acid covalently linked to the catalytic Cystein C116 | | Descriptor: | ACETATE ION, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3S3L
 
 | | Crystal Structure of CerJ from Streptomyces tendae | | Descriptor: | ACETATE ION, CHLORIDE ION, CerJ | | Authors: | Zocher, G, Hertweck, C, Bretschneider, T, Stehle, T. | | Deposit date: | 2011-05-18 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
3T6S
 
 | | Crystal structure of CerJ from Streptomyces tendae in Complex with CoA | | Descriptor: | COENZYME A, CerJ | | Authors: | Zocher, G, Bretschneider, T, Hertweck, C, Stehle, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis.
Nat.Chem.Biol., 8, 2011
|
|
