1MGV
 
 | | Crystal Structure of the R391A Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Eliot, A.C, Sandmark, J, Schneider, G, Kirsch, J.F. | | Deposit date: | 2002-08-16 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Dual-Specific Active Site of 7,8-Diaminopelargonic Acid Synthase and the Effect of the R391A Mutation
Biochemistry, 41, 2002
|
|
5OLT
 
 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Vella, P, Polyakova, A, Lindqvist, Y, Schnell, R, Bourenkov, G, Schneider, T, Schneider, G. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
5A6O
 
 | | Crystal structure of the apo form of the unphosphorylated human death associated protein kinase 3 (DAPK3) | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, S-1,2-PROPANEDIOL | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A6N
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with compound 2 | | Descriptor: | 5-(3-SULFAMOYLPHENYL)-1H-1,2,3,4-TETRAZOL-1-IDE, DEATH-ASSOCIATED PROTEIN KINASE 3, GLYCEROL, ... | | Authors: | Rodrigues, T, Reker, D, Welin, M, Caldera, M, Brunner, C, Gabernet, G, Schneider, P, Walse, B, Schneider, G. | | Deposit date: | 2015-06-30 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De Novo Fragment Design for Drug Discovery and Chemical Biology.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7JXT
 
 | | Ovine COX-1 in complex with the subtype-selective derivative 2a | | Descriptor: | 2-[4,5-bis(2-chlorophenyl)-1H-imidazol-2-yl]-6-(prop-2-en-1-yl)phenyl methoxyacetate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ko, Y, Iaselli, M, Miciaccia, M, Friedrich, L, Schneider, G, Scilimati, A, Cingolani, G. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Learning from Nature: From a Marine Natural Product to Synthetic Cyclooxygenase-1 Inhibitors by Automated De Novo Design.
Adv Sci, 8, 2021
|
|
3KOF
 
 | | Crystal structure of the double mutant F178Y/R181E of E.coli transaldolase B | | Descriptor: | SULFATE ION, Transaldolase B | | Authors: | Schneider, S, Gutierrez, M, Sandalova, T, Schneider, G, Clapes, P, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning the Active Site of Transaldolase TalB from Escherichia coli: New Variants with Improved Affinity towards Nonphosphorylated Substrates.
Chembiochem, 11, 2010
|
|
6YKD
 
 | | Human Pim-1 kinase in complex with an inhibitor identified by virtual screening | | Descriptor: | ACETATE ION, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Schneider, P, Welin, M, Svensson, B, Walse, B, Schneider, G. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Virtual Screening and Design with Machine Intelligence Applied to Pim-1 Kinase Inhibitors.
Mol Inform, 39, 2020
|
|
8R2J
 
 | |
2Y5T
 
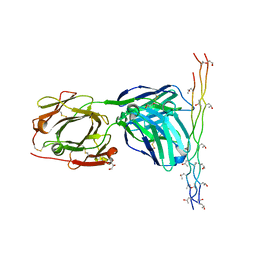 | | Crystal structure of the pathogenic autoantibody CIIC1 in complex with the triple-helical C1 peptide | | Descriptor: | C1, CHLORIDE ION, CIIC1 FAB FRAGMENT HEAVY CHAIN, ... | | Authors: | Dobritzsch, D, Lindh, I, Schneider, N, Uysal, H, Nandakumar, K.S, Burkhardt, H, Schneider, G, Holmdahl, R. | | Deposit date: | 2011-01-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Arthritogenic Anticollagen Immune Complex.
Arthritis Rheum., 63, 2011
|
|
8PBO
 
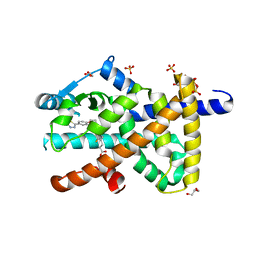 | | Deep interactome learning for generative drug design | | Descriptor: | 3-[2-fluoranyl-4-[3-[2-fluoranyl-4-(5-methyl-1,3,4-thiadiazol-2-yl)phenoxy]propoxy]phenyl]propanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Hakansson, M, Focht, D, Atz, K, Schneider, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Prospective de novo drug design with deep interactome learning.
Nat Commun, 15, 2024
|
|
8R2E
 
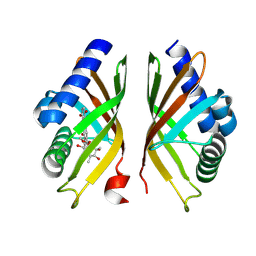 | |
8R2B
 
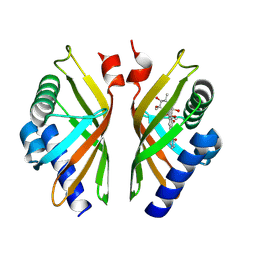 | |
8R20
 
 | |
2RUS
 
 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, MG(II), AND ACTIVATOR CO2 AT 2.3-ANGSTROMS RESOLUTION | | Descriptor: | FORMYL GROUP, MAGNESIUM ION, RUBISCO (RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE) | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1991-10-11 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ternary complex of ribulose-1,5-bisphosphate carboxylase, Mg(II), and activator CO2 at 2.3-A resolution.
Biochemistry, 30, 1991
|
|
3CWN
 
 | | Escherichia coli transaldolase b mutant f178y | | Descriptor: | SULFATE ION, Transaldolase B | | Authors: | Sandalova, T, Schneider, G, Samland, A. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Replacement of a Phenylalanine by a Tyrosine in the Active Site Confers Fructose-6-phosphate Aldolase Activity to the Transaldolase of Escherichia coli and Human Origin.
J.Biol.Chem., 283, 2008
|
|
4S2C
 
 | | Covalent complex of E. coli transaldolase TalB with fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, FRUCTOSE -6-PHOSPHATE, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1CNE
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
4S2B
 
 | | Covalent complex of E. coli transaldolase TalB with tagatose-6-phosphate | | Descriptor: | 2-deoxy-6-O-phosphono-beta-D-lyxo-hexofuranose, SULFATE ION, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2CND
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-DEPENDENT NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
2F74
 
 | | Murine MHC class I H-2Db in complex with human b2-microglobulin and LCMV-derived immunodminant peptide gp33 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Michaelsson, J, Harris, R.A, Ljunggren, H.G, Karre, K, Schneider, G, Sandalova, T. | | Deposit date: | 2005-11-30 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of the Differential Stability and Receptor Specificity of H-2D(b) in Complex with Murine versus Human beta(2)-Microglobulin.
J.Mol.Biol., 356, 2006
|
|
6EWY
 
 | | RipA Peptidoglycan hydrolase (Rv1477, Mycobacterium tuberculosis) N-terminal domain | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Schnell, R, Steiner, E.M, Schneider, G, Guy, J, Bourenkov, G. | | Deposit date: | 2017-11-07 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the N-terminal module of the cell wall hydrolase RipA and its role in regulating catalytic activity.
Proteins, 86, 2018
|
|
1I2N
 
 | |
1I2R
 
 | |
1I2P
 
 | |
1I2Q
 
 | |
