6T3L
 
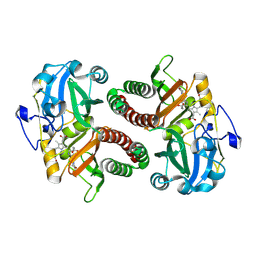 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6T3U
 
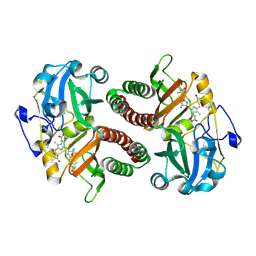 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome 1ps after photoexcitation | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6B68
 
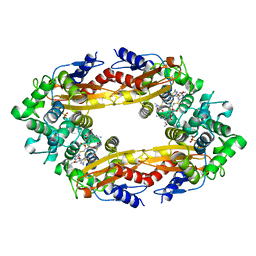 | | Beta-Lactamase, 100ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B69
 
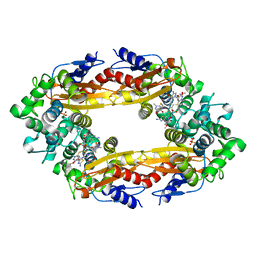 | | Beta-Lactamase, 500ms timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, (2R)-2-[(S)-{[(2E)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-(hydroxymethyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6B6D
 
 | | Beta-Lactamase, mixed with Ceftriaxone, needles crystal form, 100ms | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
6PU2
 
 | | Dark, Mutant H275T , 100K, PCM Myxobacterial Phytochrome, P2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6PTQ
 
 | | Dark, Room Temperature, PCM Myxobacterial Phytochrome, P2, Wild Type | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6PTX
 
 | | Dark, 100K, PCM Myxobacterial Phytochrome, P2, Wild Type, | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
1MD8
 
 | | Monomeric structure of the active catalytic domain of complement protease C1r | | Descriptor: | C1R COMPLEMENT SERINE PROTEASE | | Authors: | Budayova-Spano, M, Grabarse, W, Thielens, N.M, Hillen, H, Lacroix, M, Schmidt, M, Fontecilla-Camps, J, Arlaud, G.J, Gaboriaud, C. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monomeric structures of the zymogen and active catalytic domain of complement protease c1r: further insights into the c1 activation mechanism
Structure, 10, 2002
|
|
4B52
 
 | | Crystal structure of Gentlyase, the neutral metalloprotease of Paenibacillus polymyxa | | Descriptor: | BACILLOLYSIN, CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Ruf, A, Stihle, M, Benz, J, Schmidt, M, Sobek, H. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of Gentlyase, the Neutral Metalloprotease of Paenibacillus Polymyxa
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2BLI
 
 | | L29W Mb deoxy | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
7ZKY
 
 | |
2BLJ
 
 | | Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
2BLH
 
 | | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
6XLQ
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the CTX-2026 Fab | | Descriptor: | Butyrophilin subfamily 3 member A1, CTX-2026 Heavy Chain, CTX-2026 Light Chain | | Authors: | Payne, K.K, Mine, J.A, Biswas, S, Chaurio, R.A, Perales-Puchalt, A, Anadon, C.M, Costich, T.L, Harro, C.M, Walrath, J, Ming, Q, Tcyganov, E, Buras, A.L, Rigolizzo, K.E, Mandal, G, Lajoie, J, Ophir, M, Tchou, J, Marchion, D, Luca, V.C, Bobrowicz, P, McLaughlin, B, Eskiocak, U, Schmidt, M, Cubillos-Ruiz, J.R, Rodriguez, P.C, Gabrilovich, D.I, Conejo-Garcia, J.R. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BTN3A1 governs antitumor responses by coordinating alpha beta and gamma delta T cells.
Science, 369, 2020
|
|
7OVT
 
 | |
3ZRS
 
 | | X-ray crystal structure of a KirBac potassium channel highlights a mechanism of channel opening at the bundle-crossing gate. | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION | | Authors: | Bavro, V.N, De Zorzi, R, Schmidt, M.R, Muniz, J.R.C, Zubcevic, L, Sansom, M.S.P, Venien-Bryan, C, Tucker, S.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a Kirbac Potassium Channel with an Open Bundle Crossing Indicates a Mechanism of Channel Gating
Nat.Struct.Mol.Biol., 19, 2012
|
|
6ZCG
 
 | |
6ZCH
 
 | |
8B3A
 
 | |
4LP8
 
 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
6ZCF
 
 | |
7ZJ2
 
 | |
4FBM
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
8OQ4
 
 | |
