5UK0
 
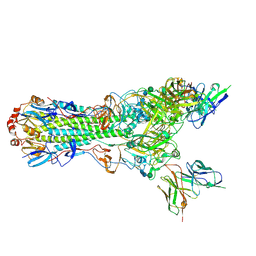 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UK1
 
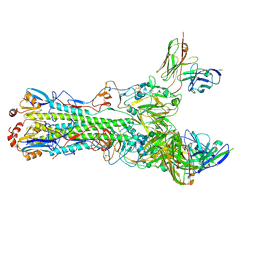 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
2LMB
 
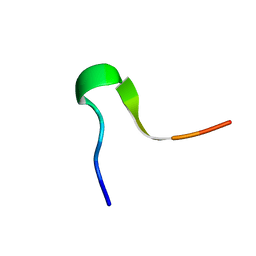 | | Solution Structure of C-terminal RAGE (ctRAGE) | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Rai, V, Maldonado, A.Y, Burz, D.S, Reverdatto, S, Schmidt, A, Shekhtman, A. | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Signal transduction in receptor for advanced glycation end products (RAGE): solution structure of C-terminal rage (ctRAGE) and its binding to mDia1.
J.Biol.Chem., 287, 2012
|
|
4J24
 
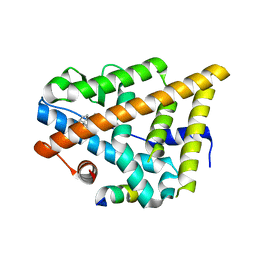 | | Estrogen Receptor in complex with proline-flanked LXXLL peptides | | Descriptor: | 19-mer peptide, ESTRADIOL, Estrogen receptor beta | | Authors: | Fuchs, S, Nguyen, H.D, Phan, T, Burton, M, Nieto, L, de Vries-van Leeuwen, I, Schmidt, A, Goodarzifard, M, Agten, S, Rose, R, Ottmann, C, Milroy, L.G, Brunsveld, L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Proline primed helix length as a modulator of the nuclear receptor-coactivator interaction
J.Am.Chem.Soc., 135, 2013
|
|
4J26
 
 | | Estrogen Receptor in complex with proline-flanked LXXLL peptides | | Descriptor: | 12-mer Peptide, ESTRADIOL, Estrogen receptor beta | | Authors: | Fuchs, S, Nguyen, H.D, Phan, T, Burton, M, Nieto, L, de Vries-van Leeuwen, I, Schmidt, A, Goodarzifard, M, Agten, S, Rose, R, Ottmann, C, Milroy, L.G, Brunsveld, L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proline primed helix length as a modulator of the nuclear receptor-coactivator interaction
J.Am.Chem.Soc., 135, 2013
|
|
4IUB
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 1 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4IUD
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form with ascorbate - partly reduced state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Hammer, M, Schmidt, A, Frielingsdorf, S, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
6G1Z
 
 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its Pfr state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Sauthof, L, Schmidt, A, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
4IUC
 
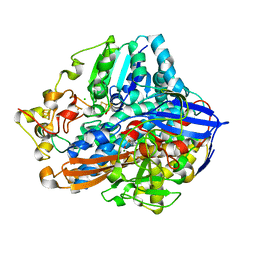 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 2 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
2J6I
 
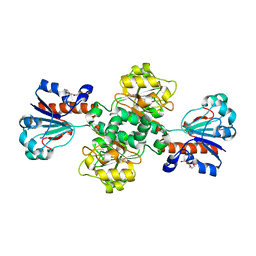 | |
6GYH
 
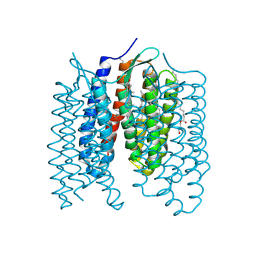 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
8AUV
 
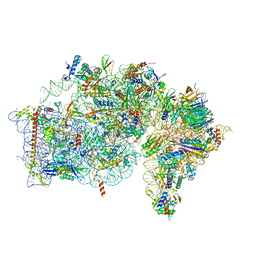 | | Cryo-EM structure of the plant 40S subunit | | Descriptor: | 18S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
5MDL
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its O2-derivatized form by a "soak-and-freeze" derivatization method | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2FSS
 
 | |
8B2L
 
 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
8AZW
 
 | | Cryo-EM structure of the plant 60S subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
2FIR
 
 | | Crystal structure of DFPR-VIIa/sTF | | Descriptor: | CALCIUM ION, CHLORIDE ION, Coagulation factor VII Heavy Chain (EC 3.4.21.21), ... | | Authors: | Bajaj, S.P, Schmidt, A.E, Padmanabhan, K, Bajaj, M.S, Prevost, D, Schreuder, H. | | Deposit date: | 2005-12-30 | | Release date: | 2006-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Structures of p-Aminobenzamidine- and Benzamidine-VIIa/Soluble Tissue Factor: Unpredicted conformation of the 192-193 peptide bond and mapping of Ca2+, Mg2+, Na+ and Zn2+ sites in factor VIIa
J.Biol.Chem., 281, 2006
|
|
7JIX
 
 | |
8POW
 
 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.61 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POX
 
 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.6 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
1BWW
 
 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QDB
 
 | | CYTOCHROME C NITRITE REDUCTASE | | Descriptor: | CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, HEME C, ... | | Authors: | Einsle, O, Messerschmidt, A, Stach, P, Huber, R, Kroneck, P.M.H. | | Deposit date: | 1999-05-19 | | Release date: | 1999-08-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of cytochrome c nitrite reductase.
Nature, 400, 1999
|
|
2UX7
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of loop shortening on the metal binding site of cupredoxin pseudoazurin.
Biochemistry, 46, 2007
|
|
2UX6
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
