4JK9
 
 | | Open and closed forms of wild-type human PRP8 RNase H-like domain with bound Co ion | | 分子名称: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | 著者 | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | 登録日 | 2013-03-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKH
 
 | | Open and closed forms of D1781E human PRP8 RNase H-like domain with bound Mg ion | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | 登録日 | 2013-03-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKC
 
 | | Open and closed forms of T1800E human PRP8 RNase H-like domain with bound Mg ion | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | 登録日 | 2013-03-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JK7
 
 | | Open and closed forms of wild-type human PRP8 RNase H-like domain with bound Mg ion | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K, MacMillan, A.M. | | 登録日 | 2013-03-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKD
 
 | | Open and closed forms of I1790Y human PRP8 RNase H-like domain with bound Mg ion | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | 著者 | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | 登録日 | 2013-03-09 | | 公開日 | 2013-05-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4J5R
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 bound to ADP-HPD | | 分子名称: | 1,2-ETHANEDIOL, 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, CHLORIDE ION, ... | | 著者 | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | 登録日 | 2013-02-09 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
1W2F
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | 分子名称: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | 著者 | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | 登録日 | 2004-07-01 | | 公開日 | 2004-09-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1W2C
 
 | | Human Inositol (1,4,5) trisphosphate 3-kinase complexed with Mn2+/AMPPNP/Ins(1,4,5)P3 | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, MANGANESE (II) ION, ... | | 著者 | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | 登録日 | 2004-07-01 | | 公開日 | 2004-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1W2D
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase complexed with Mn2+/ADP/Ins(1,3,4,5)P4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, ... | | 著者 | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | 登録日 | 2004-07-01 | | 公開日 | 2004-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | 著者 | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | 登録日 | 2022-08-05 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
6BKG
 
 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | 著者 | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | 登録日 | 2017-11-08 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
6BKF
 
 | | Lysyl-adenylate form of human LigIV catalytic domain with bound DNA substrate in open conformation | | 分子名称: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | 著者 | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | 登録日 | 2017-11-08 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
8S9K
 
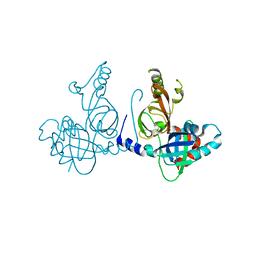 | | Structure of dimeric FAM111A SPD S541A Mutant | | 分子名称: | GLYCEROL, Serine protease FAM111A | | 著者 | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | 登録日 | 2023-03-29 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
8S9L
 
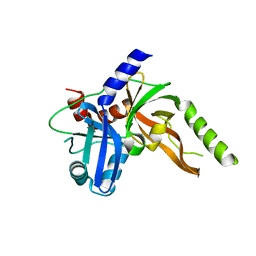 | | Structure of monomeric FAM111A SPD V347D Mutant | | 分子名称: | SULFATE ION, Serine protease FAM111A | | 著者 | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | 登録日 | 2023-03-29 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
7SAL
 
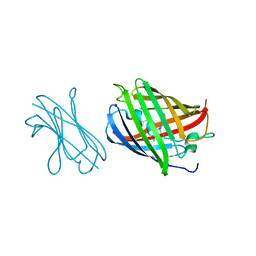 | |
7SAI
 
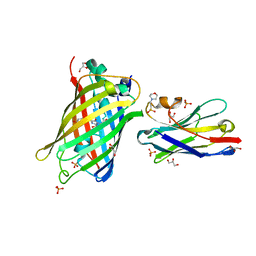 | |
7SAH
 
 | |
7SAK
 
 | |
7SAJ
 
 | |
