5G5R
 
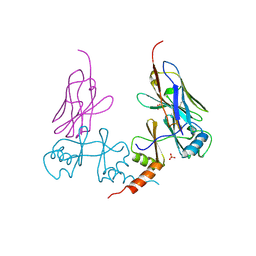 | |
5G5X
 
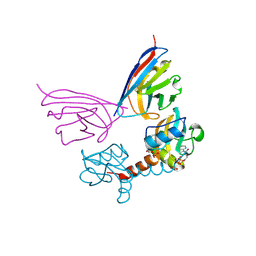 | |
8P6P
 
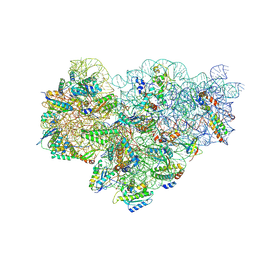 | | Mycoplasma pneumoniae small ribosomal subunit in chloramphenicol-treated cells | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-27 | | Release date: | 2024-11-20 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8P7X
 
 | | Mycoplasma pneumoniae 70S ribosome in chloramphenicol-treated cells | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-31 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8P8B
 
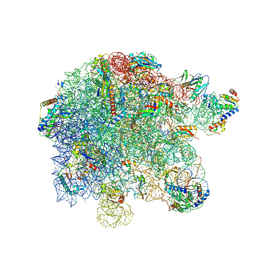 | | Mycoplasma pneumoniae large ribosomal subunit in chloramphenicol-treated cells | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-31 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8P7Y
 
 | | Mycoplasma pneumoniae 70S ribosome with second S4 protein on large subunit | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-05-31 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8P8V
 
 | | Mycoplasma pneumoniae di-ribosome in chloramphenicol-treated cells (leading 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-06-02 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8P8W
 
 | | Mycoplasma pneumoniae di-ribosome in chloramphenicol-treated cells (following 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schacherl, M, Xue, L, Spahn, C.M.T, Mahamid, J. | | Deposit date: | 2023-06-02 | | Release date: | 2024-11-20 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4BG9
 
 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (orthorhombic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BGB
 
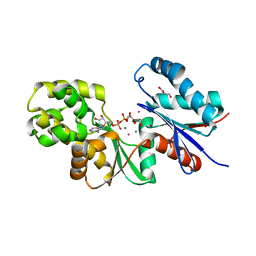 | |
4BGA
 
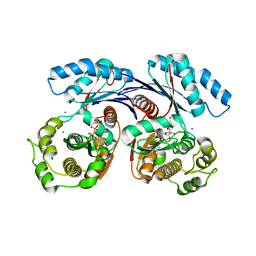 | |
4BG8
 
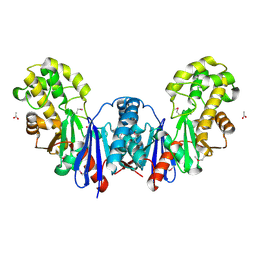 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (monoclinic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5A0P
 
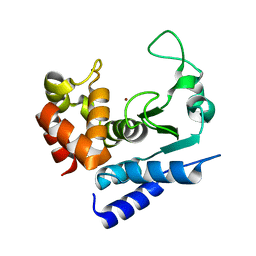 | | Apo-structure of metalloprotease Zmp1 from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0R
 
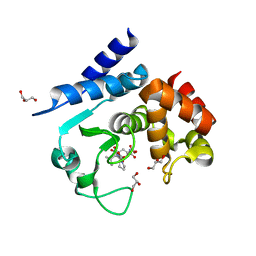 | | Product peptide-bound structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | GLYCEROL, PRODUCT PEPTIDE, ZINC ION, ... | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0S
 
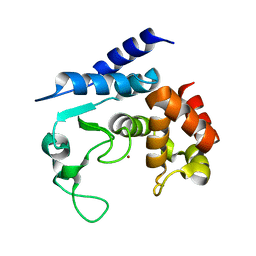 | | Apo-structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0X
 
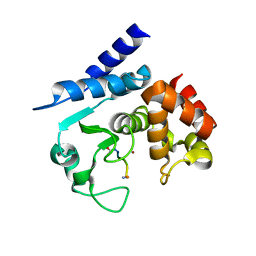 | | Substrate peptide-bound structure of metalloprotease Zmp1 variant E143AY178F from Clostridium difficile | | Descriptor: | SUBSTRATE PEPTIDE, ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
4WW0
 
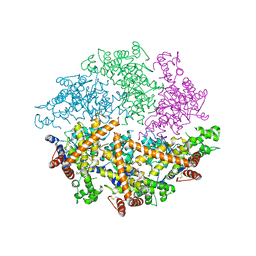 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2014-11-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FYQ
 
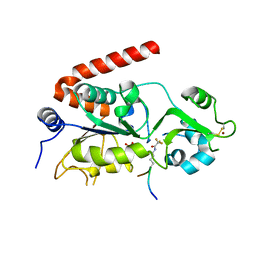 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
5N12
 
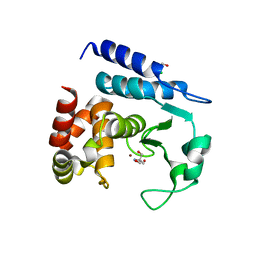 | | Crystal structure of TCE treated rPPEP-1 | | Descriptor: | 2,2,2-tris-chloroethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pro-Pro endopeptidase, ... | | Authors: | Pichlo, C, Schacherl, M, Baumann, U. | | Deposit date: | 2017-02-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
8PEG
 
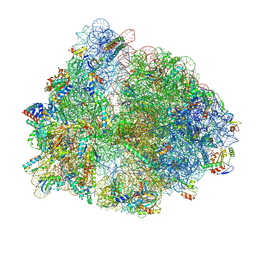 | |
8PKL
 
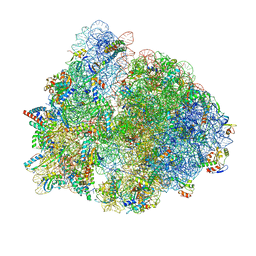 | |
5FR2
 
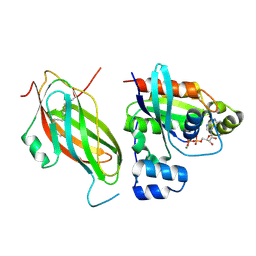 | | Farnesylated RhoA-GDP in complex with RhoGDI-alpha, lysine acetylated at K178 | | Descriptor: | FARNESYL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuhlmann, N, Wroblowski, S, Knyphausen, P, de Boor, S, Brenig, J, Zienert, A.Y, Meyer-Teschendorf, K, Praefcke, G.J.K, Nolte, H, Krueger, M, Schacherl, M, Baumann, U, James, L.C, Chin, J.W, Lammers, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural and Mechanistic Insights Into the Regulation of the Fundamental Rho-Regulator Rhogdi Alpha by Lysine Acetylation.
J.Biol.Chem., 291, 2016
|
|
5D88
 
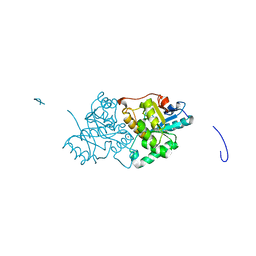 | | The Structure of the U32 Peptidase Mk0906 | | Descriptor: | ACETATE ION, Predicted protease of the collagenase family, ZINC ION | | Authors: | Baumann, U, Schacherl, M, Monatda, A.A.M. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The first crystal structure of the peptidase domain of the U32 peptidase family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6GZ2
 
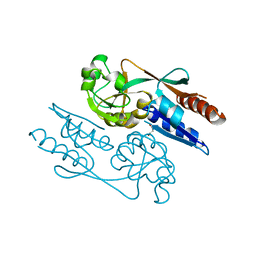 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | HTH-type transcriptional regulator LeuO, MALONATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
6GZ0
 
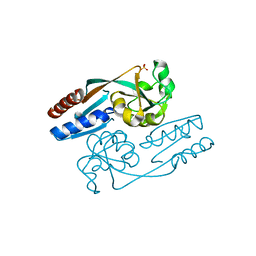 | | Crystal Structure of the LeuO Effector Binding Domain | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator LeuO, SULFATE ION | | Authors: | Fragel, S, Montada, A.M, Baumann, U, Schacherl, M, Schnetz, K. | | Deposit date: | 2018-07-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Characterization of the pleiotropic LysR-type transcription regulator LeuO of Escherichia coli.
Nucleic Acids Res., 47, 2019
|
|
