1DRW
 
 | | ESCHERICHIA COLI DHPR/NHDH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE PURIN-6-OL-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1DRU
 
 | | ESCHERICHIA COLI DHPR/NADH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1DRV
 
 | | ESCHERICHIA COLI DHPR/ACNADH COMPLEX | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1HMR
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | 9-OCTADECENOIC ACID, MUSCLE FATTY ACID BINDING PROTEIN | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1LQF
 
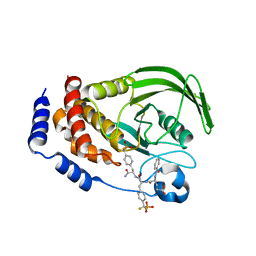 | | Structure of PTP1b in Complex with a Peptidic Bisphosphonate Inhibitor | | Descriptor: | N-BENZOYL-L-GLUTAMYL-[4-PHOSPHONO(DIFLUOROMETHYL)]-L-PHENYLALANINE-[4-PHOSPHONO(DIFLUORO-METHYL)]-L-PHENYLALANINEAMIDE, protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Asante-Appiah, E, Patel, S, Dufresne, C, Scapin, G. | | Deposit date: | 2002-05-10 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of PTP-1B in complex with a peptide inhibitor reveals an alternative binding mode for bisphosphonates.
Biochemistry, 41, 2002
|
|
7MD5
 
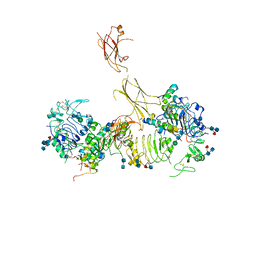 | | Insulin receptor ectodomain dimer complexed with two IRPA-9 partial agonists | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
7MD4
 
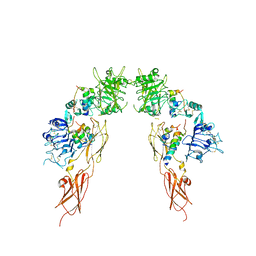 | | Insulin receptor ectodomain dimer complexed with two IRPA-3 partial agonists | | Descriptor: | Insulin B chain, Insulin chain A, Isoform Short of Insulin receptor, ... | | Authors: | Gomez-Llorente, Y, Zhou, H, Scapin, G. | | Deposit date: | 2021-04-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Nat Commun, 13, 2022
|
|
8VI4
 
 | | TehA from Haemophilus influenzae purified in LMNG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI5
 
 | | TehA from Haemophilus influenzae purified in OG | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI3
 
 | | TehA from Haemophilus influenzae purified in GDN | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VI2
 
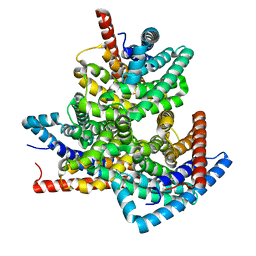 | | TehA from Haemophilus influenzae purified in DDM | | Descriptor: | Tellurite resistance protein TehA homolog | | Authors: | Catalano, C, Senko, S, Tran, N.L, Lucier, K.W, Farwell, A.C, Silva, M.S, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure Determination of Haemophilus influenzae Tellurite-Resistance Protein A via 200 kV Transmission Electron Microscopy.
Int J Mol Sci, 25, 2024
|
|
8VAE
 
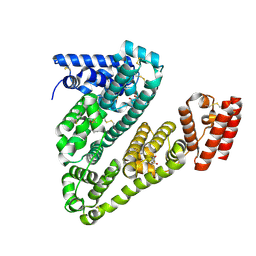 | | Cryogenic electron microscopy structure of human serum albumin in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAF
 
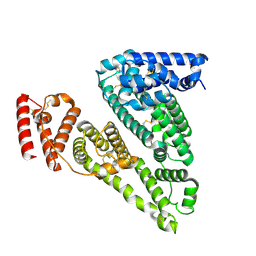 | | Cryogenic electron microscopy structure of apo human serum albumin | | Descriptor: | Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
8VAC
 
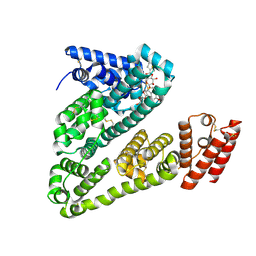 | | Cryogenic electron microscopy structure of human serum albumin in complex with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, Serum albumin | | Authors: | Catalano, C, Lucier, K.W, To, D, Senko, S, Tran, N.L, Farwell, A.C, Silva, S.M, Dip, P.V, Poweleit, N, Scapin, G. | | Deposit date: | 2023-12-11 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The CryoEM structure of human serum albumin in complex with ligands.
J.Struct.Biol., 216, 2024
|
|
7LEX
 
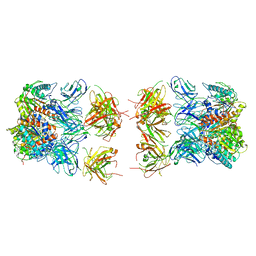 | |
7LEY
 
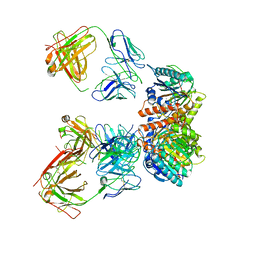 | | Trimeric human Arginase 1 in complex with mAb5 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb5 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LEZ
 
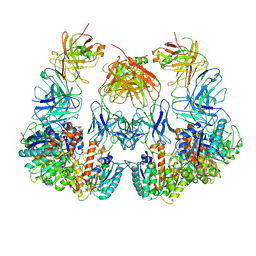 | | Trimeric human Arginase 1 in complex with mAb1 - 2 hArg:2 mAb1 complex | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb1 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF0
 
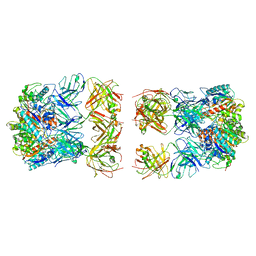 | |
7LF2
 
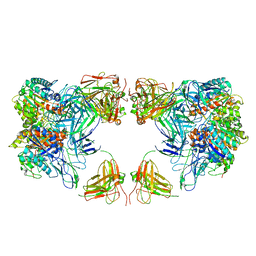 | | Trimeric human Arginase 1 in complex with mAb4 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb4 monoclonal antibody heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
7LF1
 
 | | Trimeric human Arginase 1 in complex with mAb3 | | Descriptor: | Arginase-1, MANGANESE (II) ION, mAb3 heavy chain, ... | | Authors: | Gomez-Llorente, Y, Scapin, G, Palte, R.L. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM structures of inhibitory antibodies complexed with arginase 1 provide insight into mechanism of action.
Commun Biol, 4, 2021
|
|
8UH6
 
 | | Degrader-induced complex between PTPN2 and CRBN-DDB1 | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[(4-{1-[(3R)-2,6-dioxopiperidin-3-yl]-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}piperidine-1-carbonyl)amino]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Catalano, C, Bratkowski, M, Scapin, G, Hao, Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
1R39
 
 | | THE STRUCTURE OF P38ALPHA | | Descriptor: | Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
1R3C
 
 | | THE STRUCTURE OF P38ALPHA C162S MUTANT | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
1B9H
 
 | | CRYSTAL STRUCTURE OF 3-AMINO-5-HYDROXYBENZOIC ACID (AHBA) SYNTHASE | | Descriptor: | PROTEIN (3-AMINO-5-HYDROXYBENZOIC ACID SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eads, J.C, Beeby, M, Scapin, G, Yu, T.-W, Floss, H.G. | | Deposit date: | 1999-02-11 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
Biochemistry, 38, 1999
|
|
1F06
 
 | | THREE DIMENSIONAL STRUCTURE OF THE TERNARY COMPLEX OF CORYNEBACTERIUM GLUTAMICUM DIAMINOPIMELATE DEHYDROGENASE NADPH-L-2-AMINO-6-METHYLENE-PIMELATE | | Descriptor: | L-2-AMINO-6-METHYLENE-PIMELIC ACID, MESO-DIAMINOPIMELATE D-DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cirilli, M, Scapin, G, Sutherland, A, Caplan, J.F, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 2000-05-14 | | Release date: | 2001-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-L-2-amino-6-methylene-pimelate.
Protein Sci., 9, 2000
|
|
