2IJY
 
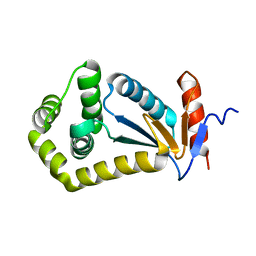 | |
1APF
 
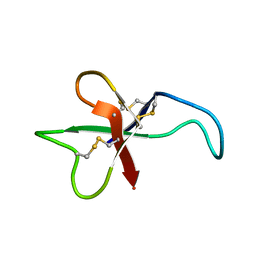 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
6BQX
 
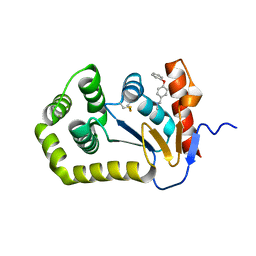 | | Crystal structure of Escherichia coli DsbA in complex with N-methyl-1-(4-phenoxyphenyl)methanamine | | Descriptor: | N-methyl-1-(4-phenoxyphenyl)methanamine, Thiol:disulfide interchange protein DsbA | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
4Z0F
 
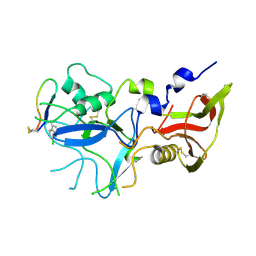 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038(6CW)] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z09
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Thr2040Ala] peptide | | Descriptor: | Apical membrane antigen 1, GLYCEROL, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0D
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038Trp] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4Z0E
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Phe2038TRN] peptide | | Descriptor: | Apical membrane antigen 1, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
4ZIJ
 
 | | Crystal structure of E.Coli DsbA in complex with 2-(4-iodophenylsulfonamido) benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Vazirani, M, Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2015-04-28 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
1AHL
 
 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-A | | Authors: | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | Deposit date: | 1994-10-28 | | Release date: | 1995-11-14 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
6BR4
 
 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
6VUB
 
 | |
6VUF
 
 | |
6VUC
 
 | |
6VUJ
 
 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 15c (N,N-diethyl-3',4'-dimethoxy-6-(1-methyl-5-oxopyrrolidin-3-yl)-[1,1'-biphenyl]-3-sulfonamide) | | Descriptor: | Bromodomain-containing protein 4, N,N-diethyl-3',4'-dimethoxy-6-[(3S)-1-methyl-5-oxopyrrolidin-3-yl][1,1'-biphenyl]-3-sulfonamide, NITRATE ION | | Authors: | Ilyichova, O.V, Scanlon, M.J, Thompson, P.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substituted 1-methyl-4-phenylpyrrolidin-2-ones - Fragment-based design of N-methylpyrrolidone-derived bromodomain inhibitors.
Eur.J.Med.Chem., 191, 2020
|
|
7LHP
 
 | |
2MBT
 
 | | NMR study of PaDsbA | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Rimmer, K, Mohanty, B, Scanlon, M.J. | | Deposit date: | 2013-08-03 | | Release date: | 2014-11-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
2NDO
 
 | | Structure of EcDsbA-sulfonamide1 complex | | Descriptor: | 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Williams, M.L, Doak, B.C, Vazirani, M, Ilyichova, O, Wang, G, Bermel, W, Simpson, J.S, Chalmers, D.K, King, G.F, Mobli, M, Scanlon, M.J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
2MBS
 
 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
2ZNM
 
 | | Oxidoreductase NmDsbA3 from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Robertson, A.L, Bottomley, S.P, Horne, J, Chin, Y, Velkov, T, Wielens, J, Thompson, P.E, Piek, S, Byres, E, Beddoe, T, Wilce, M.C.J, Kahler, C, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Oxidoreductase NmDsbA3 from Neisseria meningitidis
J.Biol.Chem., 283, 2008
|
|
3AKM
 
 | | X-ray structure of iFABP from human and rat with bound fluorescent fatty acid analogue | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, Fatty acid-binding protein, intestinal, ... | | Authors: | Wielens, J, Laguerre, A.J.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-07-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human and rat intestinal fatty acid binding proteins in complex with 11-(Dansylamino)undecanoic acid
To be Published
|
|
3AKN
 
 | |
3AO1
 
 | | Fragment-based approach to the design of ligands targeting a novel site in HIV-1 integrase | | Descriptor: | 1,3-benzodioxol-5-ol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO5
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 5-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-3-amine, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO2
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3A3T
 
 | | The oxidoreductase NmDsbA1 from N. meningitidis | | Descriptor: | Thiol:disulfide interchange protein DsbA | | Authors: | Vivian, J.P, Scoullar, J, Bushell, S, Beddoe, T, Wilce, M.C.J, Byres, E, Boyle, T.P, Rimmer, K, Doak, B, Simpson, J.S, Graham, B, Heras, B, Kahler, C.M, Rossjohn, J, Scanlon, M.J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis
J.Mol.Biol., 394, 2009
|
|
