8FXQ
 
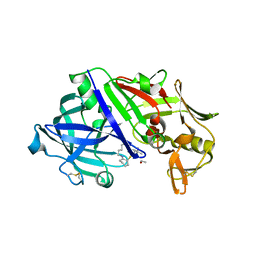 | | The Crystal Sturucture of Rhizopuspepsin with a bound modified peptide inhibitor generated by de novo drug design. | | Descriptor: | ALA-CYS-VAL-LYS, CYCLOHEXANE, Rhizopuspepsin, ... | | Authors: | Satyshur, K.A, Rich, D.H, Ripka, A.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Aspartic protease inhibitors designed from computer-generated templates bind as predicted.
Org Lett, 3, 2001
|
|
6D9Q
 
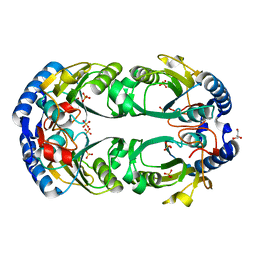 | | The sulfate-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | GLYCEROL, Hypoxanthine phosphoribosyltransferase, SULFATE ION | | Authors: | Satyshur, K.A, Dubiel, K, Anderson, B, Wolak, C, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
6DCR
 
 | |
6DGD
 
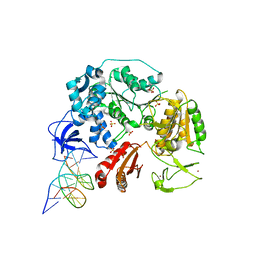 | | PriA helicase bound to dsDNA of a DNA replication fork | | Descriptor: | DNA (5'-D(P*AP*GP*CP*AP*CP*GP*CP*CP*GP*AP*CP*T)-3'), DNA (5'-D(P*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*AP*CP*T)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Satyshur, K.A, Windgassen, T.A, Keck, J.L. | | Deposit date: | 2018-05-17 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.823 Å) | | Cite: | Structure-specific DNA replication-fork recognition directs helicase and replication restart activities of the PriA helicase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2EWV
 
 | |
2EYU
 
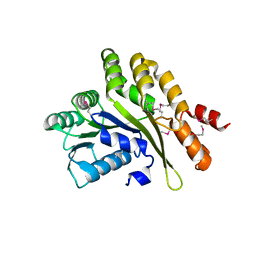 | | The Crystal Structure of the C-terminal Domain of Aquifex aeolicus PilT | | Descriptor: | SULFATE ION, twitching motility protein PilT | | Authors: | Satyshur, K.A, Worzalla, G.A, Meyer, L.S, Heiniger, E.K, Aukema, K.G, Forest, K.T. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility.
Structure, 15, 2007
|
|
2EWW
 
 | |
6VBT
 
 | |
6VBS
 
 | |
6W1I
 
 | | Re-interpretation of ppGpp (G4P) electron density in the deposited crystal structure of Xanthine phosphoribosyltransferase (XPRT) (1Y0B). | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, SODIUM ION, Xanthine phosphoribosyltransferase | | Authors: | Satyshur, K.A, Anderson, B.W, Keck, J.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of Regulation of the Purine Salvage Enzyme XPRT by the Alarmones pppGpp, ppGpp, and pGpp.
J.Mol.Biol., 432, 2020
|
|
6D9R
 
 | | The substrate-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | 1,2-ETHANEDIOL, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAGUANINE, ... | | Authors: | Satyshur, K.A, Wolak, C, Anderson, B, Dubiel, K, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
6D9S
 
 | | The (p)ppGpp-bound crystal structure of HPRT (hypoxanthine phosphoribosyltransferase) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5',3'-TETRAPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Satyshur, K.A, Dubiel, K, Anderson, B, Wolak, C, Keck, J.L. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Evolution of (p)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction.
Elife, 8, 2019
|
|
3HL8
 
 | | Crystal structure of exonuclease I in complex with inhibitor BCBP | | Descriptor: | (5R)-3-tert-butyl-1-(6-chloro-1,3-benzothiazol-2-yl)-4,5-dihydro-1H-pyrazol-5-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Satyshur, K.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HP9
 
 | | Crystal structure of SSB/Exonuclease I in complex with inhibitor CFAM | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-chloro-5-(trifluoromethyl)phenyl]amino}-5-methoxybenzoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Satyshur, K.A. | | Deposit date: | 2009-06-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IPN
 
 | | Crystal Structure of fluorine and methyl modified collagen: (mepFlpgly)7 | | Descriptor: | CARBONATE ION, Non-natural Collagen | | Authors: | Satyshur, K.A, Shoulders, M.D, Raines, R.T, Forest, K.T. | | Deposit date: | 2009-08-18 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Stereoelectronic and steric effects in side chains preorganize a protein main chain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MXN
 
 | | Crystal structure of the RMI core complex | | Descriptor: | BENZAMIDINE, RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Hoadley, K.A, Xu, D, Xue, Y, Satyshur, K.A, Wang, W, Keck, J.L. | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome.
Structure, 18, 2010
|
|
4Z1W
 
 | | CRYSTAL STRUCTURE OF MONOMERIC BACTERIOPHYTOCHROME mutant D207L Y263F From Synchrotron | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Bhattacharya, S, Satyshur, K.A, Wangkanont, K, Lehtivuori, H, Forest, K.T. | | Deposit date: | 2015-03-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor.
Front Mol Biosci, 2, 2015
|
|
7R7J
 
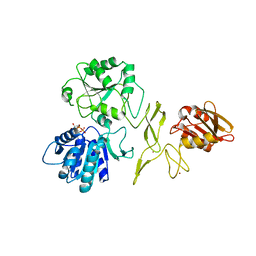 | | Crystal structure of RadD with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative DNA repair helicase RadD, ... | | Authors: | Osorio Garcia, M.A, Satyshur, K.A, Keck, J.L, Cox, M.M. | | Deposit date: | 2021-06-24 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the Escherichia coli RadD DNA repair protein bound to ADP reveals a novel zinc ribbon domain.
Plos One, 17, 2022
|
|
5IC5
 
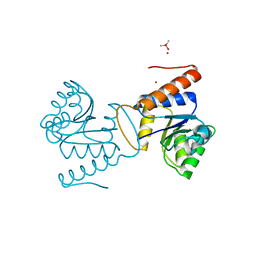 | | Bacteriophytochrome response regulator RtBRR | | Descriptor: | CACODYLATE ION, Candidate response regulator, CheY, ... | | Authors: | Baker, A.W, Satyshur, K.A, Forest, K.T. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arm-in-Arm Response Regulator Dimers Promote Intermolecular Signal Transduction.
J.Bacteriol., 198, 2016
|
|
6NQC
 
 | |
6NIV
 
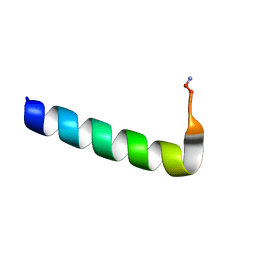 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
4LAC
 
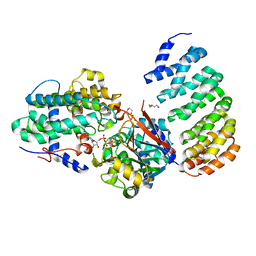 | | Crystal Structure of Protein Phosphatase 2A (PP2A) and PP2A phosphatase activator (PTPA) complex with ATPgammaS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Guo, F, Stanevich, V, Wlodarchak, N, Satyshur, K.A, Xing, Y. | | Deposit date: | 2013-06-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone.
Cell Res., 24, 2014
|
|
3NCT
 
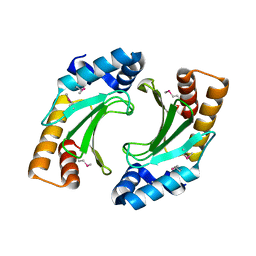 | | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of reca | | Descriptor: | Protein psiB | | Authors: | Petrova, V, Satyshur, K.A, George, N.P, McCaslin, D, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of RecA.
J.Biol.Chem., 285, 2010
|
|
3P71
 
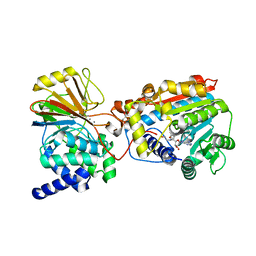 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
8FAK
 
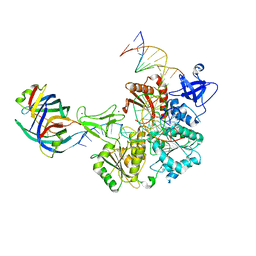 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
