3JVV
 
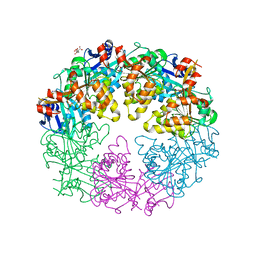 | | Crystal Structure of P. aeruginosa PilT with bound AMP-PCP | | Descriptor: | CITRIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Misic, A.M, Satyshur, K.A, Forest, K.T. | | Deposit date: | 2009-09-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor.
J.Mol.Biol., 400, 2010
|
|
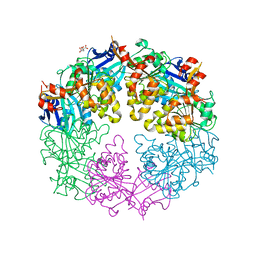
3JVU
 
 | |
3NCT
 
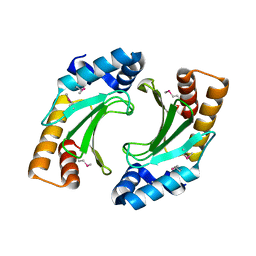 | | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of reca | | Descriptor: | Protein psiB | | Authors: | Petrova, V, Satyshur, K.A, George, N.P, McCaslin, D, Cox, M.M, Keck, J.L. | | Deposit date: | 2010-06-05 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the bacterial conjugation factor PsiB, a negative regulator of RecA.
J.Biol.Chem., 285, 2010
|
|
3P71
 
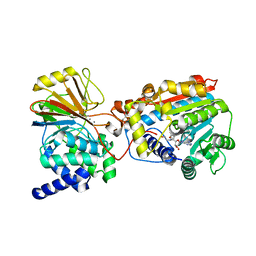 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
4ZRR
 
 | | Crystal Structure of Monomeric Bacteriophytochrome mutant D207L Y263F at 1.5 A resolution Using a home source. | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Bhattacharya, S, Satyshur, K.A, Lehtivuori, H, Forest, K.T. | | Deposit date: | 2015-05-12 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor.
Front Mol Biosci, 2, 2015
|
|
5BRJ
 
 | |
5CGN
 
 | | Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 8 | | Descriptor: | CHLORIDE ION, D-Ala-Magainin Derivative, L-ACPC8-Ala-Magainin | | Authors: | Hayouka, Z, Thomas, N.C, Mortenson, D.E, Satyshur, K.A, Weisblum, B, Forest, K.T, Gellman, S.H. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quasiracemate Crystal Structures of Magainin 2 Derivatives Support the Functional Significance of the Phenylalanine Zipper Motif.
J.Am.Chem.Soc., 137, 2015
|
|
5CGO
 
 | | Structure of quasiracemic Ala-Magainin 2 with a beta amino acid substitution at position 13 | | Descriptor: | ACPC-13 derivative of Ala-Magainin 2, D-Ala-Magainin 2 | | Authors: | Hayouka, Z, Thomas, N.C, Mortenson, D.E, Satyshur, K.A, Weisblum, B, Forest, K.T, Gellman, S.H. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Quasiracemate Crystal Structures of Magainin 2 Derivatives Support the Functional Significance of the Phenylalanine Zipper Motif.
J.Am.Chem.Soc., 137, 2015
|
|
2GSZ
 
 | |
6D52
 
 | |
4H5B
 
 | | Crystal Structure of DR_1245 from Deinococcus radiodurans | | Descriptor: | BROMIDE ION, DR_1245 protein, GLYCEROL, ... | | Authors: | Norais, C, Servant, P, Bouthier-de-la-Tour, C, Coureux, P.D, Ithurbide, S, Vannier, F, Guerin, P, Dulberger, C.L, Satyshur, K.A, Keck, J.L, Armengaud, J, Cox, M.M, Sommer, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Deinococcus radiodurans DR1245 Protein, a DdrB Partner Homologous to YbjN Proteins and Reminiscent of Type III Secretion System Chaperones.
Plos One, 8, 2013
|
|
4HJD
 
 | | GCN4pLI derivative with alpha/beta/acyclic-gamma amino acid substitution pattern | | Descriptor: | GCN4pLI(alpha/beta/acyclic gamma) | | Authors: | Shin, Y.H, Mortenson, D.E, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Differential Impact of beta and gamma Residue Preorganization on alpha / beta / gamma-Peptide Helix Stability in Water.
J.Am.Chem.Soc., 135, 2013
|
|
4I5N
 
 | | Structural mechanism of trimeric PP2A holoenzyme involving PR70: insight for Cdc6 dephosphorylation | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Microcystin-LR (MCLR) bound form, ... | | Authors: | Wlodarchak, N, Satyshur, K.A, Guo, F, Xing, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
4HJB
 
 | | GCN4pLI derivative with alpha/beta/cyclic-gamma amino acid substitution pattern | | Descriptor: | GCN4pLI(alpha/beta/cyclic-gamma) | | Authors: | Shin, Y.H, Mortenson, D.E, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2012-10-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Differential Impact of beta and gamma Residue Preorganization on alpha / beta / gamma-Peptide Helix Stability in Water.
J.Am.Chem.Soc., 135, 2013
|
|
4I5L
 
 | | Structural mechanism of trimeric PP2A holoenzyme involving PR70: insight for Cdc6 dephosphorylation | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Wlodarchak, N, Satyshur, K.A, Guo, F, Xing, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
3S7Q
 
 | | Crystal Structure of a Monomeric Infrared Fluorescent Deinococcus radiodurans Bacteriophytochrome chromophore binding domain | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Auldridge, M.E, Satyshur, K.A, Forest, K.T. | | Deposit date: | 2011-05-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein.
J.Biol.Chem., 287, 2012
|
|
3S7N
 
 | |
3S7O
 
 | | Crystal Structure of the Infrared Fluorescent D207H variant of Deinococcus Bacteriophytochrome chromophore binding domain at 1.24 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, GLYCEROL | | Authors: | Forest, K.T, Auldridge, M.E, Satyshur, K.A, Anstrom, D.M. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein.
J.Biol.Chem., 287, 2012
|
|
3S7P
 
 | |
3TRW
 
 | |
3TJW
 
 | | Crystal Structure of Quasiracemic Villin Headpiece Subdomain Containing (F5Phe10) Substitution | | Descriptor: | D-Villin-1, L-Villin-1, SULFATE ION | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-01-25 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
3TRV
 
 | | Crystal structure of quasiracemic villin headpiece subdomain containing (F5Phe17) substitution | | Descriptor: | D-Villin-1, ISOPROPYL ALCOHOL, L-Villin-1, ... | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
3TRY
 
 | | Crystal structure of racemic villin headpiece subdomain in space group I-4c2 | | Descriptor: | D-Villin-1, SULFATE ION | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-09-11 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
6BHX
 
 | | B. subtilis SsbA with DNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein A | | Authors: | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | Deposit date: | 2017-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.936 Å) | | Cite: | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
6BHW
 
 | | B. subtilis SsbA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Single-stranded DNA-binding protein A | | Authors: | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | Deposit date: | 2017-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
