4QI2
 
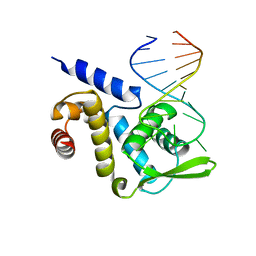 | | X-ray structure of the ROQ domain from murine Roquin-1 in complex with a 23-mer Tnf-CDE RNA | | Descriptor: | RNA (5'-R(*AP*CP*AP*UP*GP*UP*UP*UP*UP*CP*UP*GP*UP*GP*AP*AP*AP*AP*CP*GP*GP*AP*G)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4JA7
 
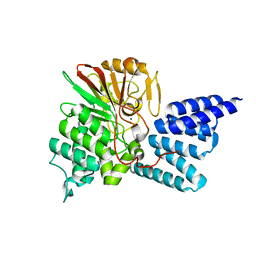 | | Rat PP5 co-crystallized with P5SA-2 | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
2W4S
 
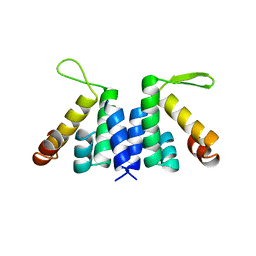 | | novel RNA-binding domain in Cryptosporidium parvum at 2.5 angstrom resolution | | Descriptor: | Ankyrin-repeat protein | | Authors: | Varrot, A, Mackereth, C, Mourao, A, Sattler, M, Cusack, S. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and RNA Recognition by the Snrna and Snorna Transport Factor Phax.
RNA, 16, 2010
|
|
4BA8
 
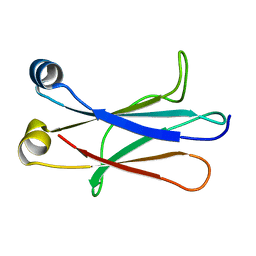 | | High resolution NMR structure of the C mu3 domain from IgM | | Descriptor: | IG MU CHAIN C REGION SECRETED FORM | | Authors: | Mueller, R, Kern, T, Graewert, M.A, Madl, T, Peschek, J, Groll, M, Sattler, M, Buchner, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-06-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High Resolution Structures of the Igm Fc Domains Reveal Principles of its Hexamer Formation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2L1L
 
 | |
1ZZJ
 
 | | Structure of the third KH domain of hnRNP K in complex with 15-mer ssDNA | | Descriptor: | 5'-D(*TP*TP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*TP*TP*T)-3', Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
1ZZK
 
 | | Crystal Structure of the third KH domain of hnRNP K at 0.95A resolution | | Descriptor: | Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
6R78
 
 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R79
 
 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop open) | | Descriptor: | BETA-MERCAPTOETHANOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
5MMC
 
 | |
6RZS
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed ertapenem | | Descriptor: | Beta-lactamase, ZINC ION, hydrolysed ertapenem | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
1OPI
 
 | | SOLUTION STRUCTURE OF THE THIRD RNA RECOGNITION MOTIF (RRM) OF U2AF65 IN COMPLEX WITH AN N-TERMINAL SF1 PEPTIDE | | Descriptor: | SPLICING FACTOR SF1, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
6RZR
 
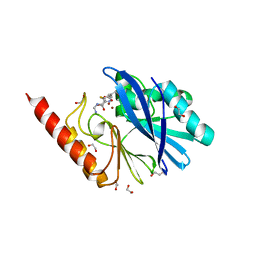 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed imipenem | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
4BXU
 
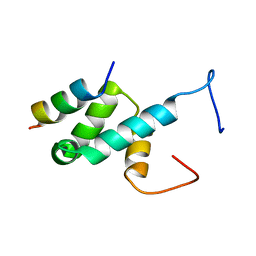 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
6SPT
 
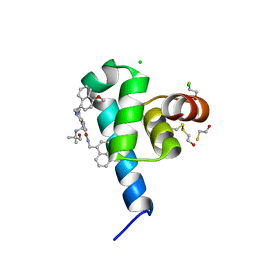 | | High resolution crystal structure of N-terminal domain of PEX14 from Trypanosoma brucei in complex with the fist compound with sub-micromolar trypanocidal activity | | Descriptor: | 5-[(4-methoxynaphthalen-1-yl)methyl]-1-[2-[(2-methyl-1-oxidanyl-propan-2-yl)amino]ethyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Napolitano, V, Dawidowski, M, Kalel, V.C, Fino, R, Emmanouilidis, L, Lenhart, D, Ostertag, M, Kaiser, M, Kolonko, M, Schilebs, W, Maser, P, Tetko, I, Hadian, K, Plettenburg, O, Erdmann, R, Sattler, M, Popowicz, G.M, Dubin, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Activity Relationship in Pyrazolo[4,3-c]pyridines, First Inhibitors of PEX14-PEX5 Protein-Protein Interaction with Trypanocidal Activity.
J.Med.Chem., 63, 2020
|
|
5N8V
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1-(2-azanylethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
4B9W
 
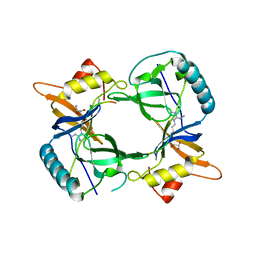 | | Structure of extended Tudor domain TD3 from mouse TDRD1 in complex with MILI peptide containing dimethylarginine 45. | | Descriptor: | GLYCEROL, PIWI-LIKE PROTEIN 2, TUDOR DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Mathioudakis, N, Palencia, A, Kadlec, J, Round, A, Tripsianes, K, Sattler, M, Pillai, R.S, Cusack, S. | | Deposit date: | 2012-09-08 | | Release date: | 2012-10-17 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.
Rna, 18, 2012
|
|
6S0H
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed doripenem | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-2,3-dihydro-1~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
4A4E
 
 | | Solution structure of SMN Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL MOTOR NEURON PROTEIN | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4H
 
 | | Solution structure of SPF30 Tudor domain in complex with asymmetrically dimethylated arginine | | Descriptor: | NG,NG-DIMETHYL-L-ARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A4F
 
 | | Solution structure of SPF30 Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
1LXL
 
 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
2XNF
 
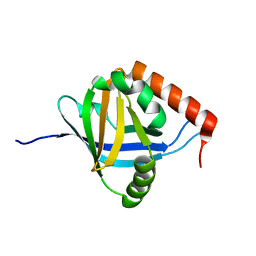 | | The Mediator Med25 activator interaction domain: Structure and cooperative binding of VP16 subdomains | | Descriptor: | MEDIATOR OF RNA POLYMERASE II TRANSCRIPTION SUBUNIT 25 | | Authors: | Vojnic, E, Mourao, A, Seizl, M, Simon, B, Wenzeck, L, Lariviere, L, Baumli, S, Meisterernst, M, Sattler, M, Cramer, P. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Vp16 Binding of the Mediator Med25 Activator Interaction Domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4A24
 
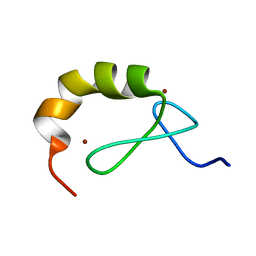 | | Structural and functional analysis of the DEAF-1 and BS69 MYND domains | | Descriptor: | DEFORMED EPIDERMAL AUTOREGULATORY FACTOR 1 HOMOLOG, ZINC ION | | Authors: | Kateb, F, Perrin, H, Tripsianes, K, Zou, P, Spadaccini, R, Bottomley, M, Bepperling, A, Ansieau, S, Sattler, M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of the Deaf-1 and Bs69 Mynd Domains.
Plos One, 8, 2013
|
|
6R8G
 
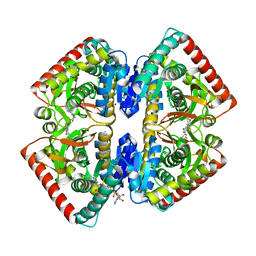 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with 4-(3,4-difluorophenyl)thiazol-2-amine | | Descriptor: | 4-[3,4-bis(fluoranyl)phenyl]-1,3-thiazol-2-amine, Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fragment-Based Approach Identifies an Allosteric Pocket that Impacts Malate Dehydrogenase Activity
Commun Biol, 2021
|
|
